Microspectrophotometry in Marine Visual Ecology: Techniques, Applications, and Advances in Drug Discovery
This article provides a comprehensive overview of microspectrophotometry (MSP) as a pivotal technique for analyzing visual pigments in marine species.
Microspectrophotometry in Marine Visual Ecology: Techniques, Applications, and Advances in Drug Discovery
Abstract
This article provides a comprehensive overview of microspectrophotometry (MSP) as a pivotal technique for analyzing visual pigments in marine species. Tailored for researchers, scientists, and drug development professionals, it covers the foundational principles of MSP, detailed methodological protocols for marine applications, and strategies for troubleshooting and data validation. By exploring its use in diverse organisms—from deep-sea crustaceans to flatfish—the content highlights how MSP-derived insights into spectral sensitivity and visual adaptation are informing the discovery of novel bioactive compounds and visual system models with biomedical potential.
Unlocking Marine Vision: The Foundational Role of Microspectrophotometry
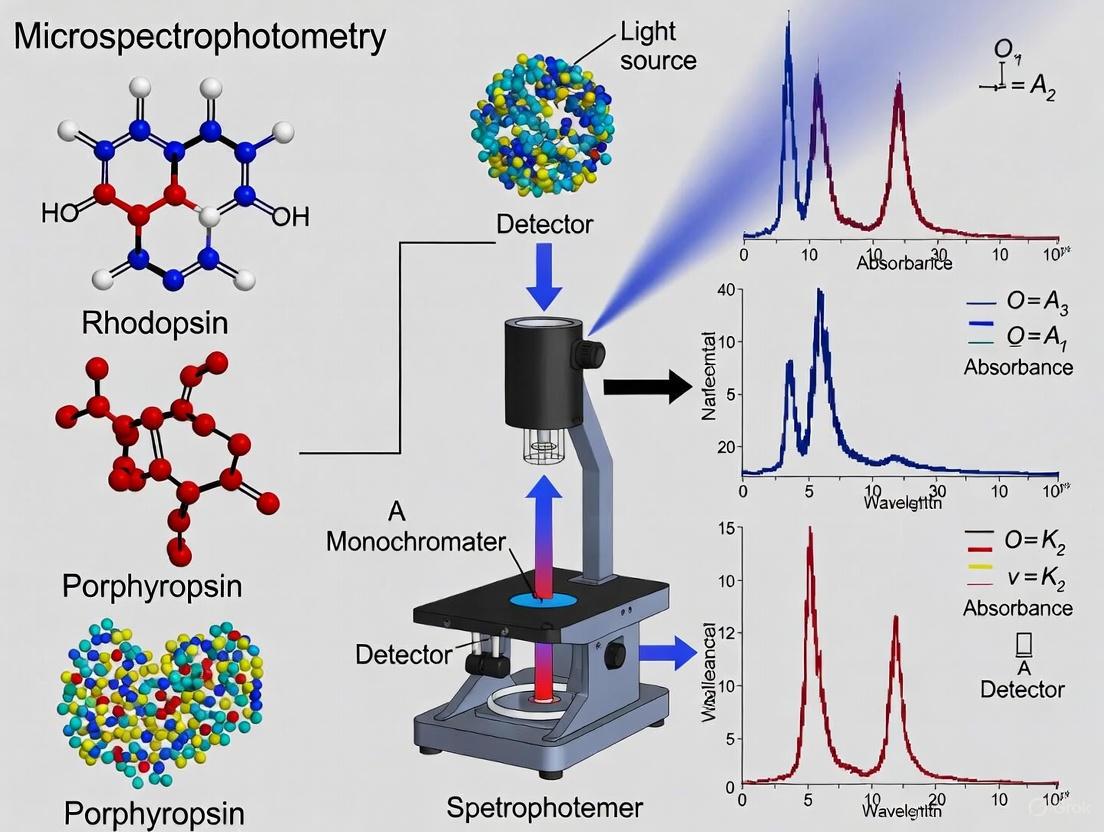
Microspectrophotometry (MSP) is a cornerstone technique in visual neuroscience for directly measuring the absorbance properties of visual pigments within individual photoreceptor cells. This method is particularly vital for studying marine species, whose visual systems have evolved complex adaptations to the unique light conditions of aquatic environments. By enabling the analysis of single cells, MSP provides researchers with precise, cell-specific data on spectral sensitivity, revealing the chromatic basis of vision in everything from deep-sea fish to coastal species.
The Core Measurement Principle of MSP
Fundamental Operating Principle
The fundamental principle of MSP is to pass a beam of monochromatic light through a single, isolated photoreceptor cell and measure the proportion of light that is absorbed at each wavelength. Visual pigments, which are housed in the outer segments of photoreceptor cells, have characteristic bell-shaped absorption spectra [1]. The key parameter derived from an MSP measurement is the wavelength of maximum absorption (λmax), which denotes the specific wavelength at which the visual pigment absorbs the most light and is a direct indicator of the pigment's spectral sensitivity [1].
The measurement is based on the Beer-Lambert law, which relates the absorption of light to the properties of the material through which the light is traveling. A higher density of visual pigment molecules in the light path results in greater light absorption. The ratio of transmitted light through the cell to a reference beam (often through a blank area of the preparation) is calculated across the spectrum, yielding a transverse absorbance spectrum for that single cell.
The following diagram illustrates the generalized workflow for an MSP experiment, from sample preparation to data analysis.
Detailed Experimental Protocol
Sample Preparation and Mounting
- Tissue Extraction: Dark-adapt the animal for at least one hour prior to dissection to allow visual pigments to regenerate fully. Under dim, deep-red light, enucleate the eye and hemisect it to remove the retina.
- Photoreceptor Isolation: Place the retinal piece in a droplet of appropriate physiological saline or buffer on a microscope coverslip. Gently macerate the tissue using fine needles or a small vibrator to liberate individual photoreceptor cells.
- Slide Preparation: Introduce a small volume of glycerol-based mounting medium to the suspension to improve optical clarity. Carefully lower a glass microscope slide onto the coverslip to create a sealed preparation. The sample should be kept cool and dark until measurement.
Instrumentation and Data Acquisition
- Microscope Setup: MSP is performed on a modified microscope that combines a high-quality optical microscope with a scanning monochromator and a sensitive photodetector (e.g., a photomultiplier tube or CCD).
- Cell Selection and Alignment: View the sample under infrared light using an infrared-sensitive camera to avoid bleaching the visual pigments. Select an intact photoreceptor and meticulously align its outer segment so it is perfectly positioned in the path of the measuring beam.
- Spectral Scanning: Scan through a range of wavelengths (e.g., from 350 nm to 700 nm), measuring the intensity of light transmitted through the cell (I) and through a reference blank area next to the cell (I~0~) at each wavelength step.
- Bleaching Control: Expose the cell to intense white light for several minutes to photobleach the visual pigment. Repeat the spectral scan to obtain a "bleached" baseline spectrum.
Data Processing and Analysis
- Absorbance Calculation: For each wavelength, compute the absorbance (A) using the formula: ( A = \log{10} (I0 / I) ) This generates a raw absorbance spectrum.
- Baseline Correction: Subtract the bleached spectrum from the raw absorbance spectrum to obtain a difference spectrum, which represents the specific absorbance of the visual pigment.
- Normalization and Curve Fitting: Normalize the difference spectrum to its peak absorbance. Fit a standard visual pigment template curve (e.g., a Govardovskii or Dartnall template) to the data points to determine the precise λmax [2] [3].
Key Data and Applications in Marine Research
Representative MSP Data from Marine Fish
MSP has revealed a remarkable diversity of visual pigments in marine fishes, adapted to their specific photic environments. The table below summarizes example λmax values measured in various species, illustrating this diversity.
Table 1: Visual Pigment λmax Values in Selected Marine Fishes
| Species | Common Name | Photoreceptor Type | λmax (nm) | Citation |
|---|---|---|---|---|
| Hippoglossus hippoglossus | Atlantic Halibut | Rod | 491 | [2] |
| Single Cone (S) | 431, 457 | [2] | ||
| Single Cone (M) | 500, 514, 527 | [2] | ||
| Single Cone (L) | 550 | [2] | ||
| Mugil cephalus | Flathead Grey Mullet | Rod | 506 | [3] |
| Single Cone (S) | 464 | [3] | ||
| Single Cone (M) | 518 | [3] | ||
| Double Cone (L1/L2) | 560, 574 | [3] | ||
| Pleuronectes americanus | Winter Flounder | Pre-metamorphosis Cone | 519 | [1] |
| Post-metamorphosis Single Cone | 457 | [1] | ||
| Post-metamorphosis Double Cone | 531, 547 | [1] |
These measurements are crucial for understanding visual ecology. For instance, the spectral shift in the rod pigments of deep-sea fish (λmax ~470-490 nm) is an adaptation to the dominant blue light of the deep ocean and the spectrum of bioluminescent emissions [1].
Integrating MSP with Other Techniques
MSP is often used in conjunction with other methods to build a comprehensive understanding of visual systems. As exemplified in a 2022 study on Atlantic halibut, MSP can be correlated with ophthalmic histology to connect spectral sensitivity with retinal structure, and with bioinformatic analysis of the opsin gene repertoire to link functional pigment expression with genetic potential [2]. Another powerful combination is the use of liquid chromatography, as seen in mullet studies, to determine the specific chromophore (e.g., A1- vs. A2-based) used in the visual pigments, which can further tune their λmax [3].
The conceptual relationships between MSP and these complementary disciplines are shown below.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful MSP experimentation relies on a suite of specialized reagents and instruments. The following table details key items and their functions in the protocol.
Table 2: Essential Reagents and Materials for MSP
| Item/Category | Specific Examples & Details | Critical Function in Protocol |
|---|---|---|
| Physiological Saline | Teleost Ringer's solution, phosphate-buffered saline (PBS). | Maintains osmotic balance and cellular integrity of photoreceptors during dissection and isolation. |
| Mounting Medium | Glycerol-based solutions (e.g., 50-90% glycerol in buffer). | Provides an optically clear medium that minimizes light scattering for accurate absorbance measurements. |
| Protease Inhibitors | Cocktails (e.g., containing AEBSF, EDTA). | Added to saline to slow proteolytic degradation of visual pigments and outer segments. |
| Microscope & Optics | UV-VIS capable microscope, high-NA objectives, IR light source. | Forms the core platform for visualizing cells (via IR) and delivering/collecting measuring light. |
| Monochromator | Scanning grating monochromator. | Generates a highly pure, wavelength-specific beam of light for the spectral scan. |
| Photodetector | Photomultiplier Tube (PMT), sensitive CCD camera. | Precisely measures the intensity of light transmitted through the single cell. |
The marine environment presents unique challenges for vision, leading to the evolution of highly specialized visual systems. Research into the anatomy of marine vision has revealed remarkable adaptations in photoreceptors, retinal mosaics, and optical structures across diverse species. These adaptations are crucial for survival in various photic niches, from surface waters to the deep sea. Microspectrophotometry (MSP) has emerged as a pivotal technique for investigating visual pigments in marine organisms, allowing researchers to measure absorbance spectra of individual photoreceptor cells and correlate them with molecular genetics and ecological adaptations [2] [4] [5].
The significance of this field extends beyond basic biological understanding, with applications in drug development where marine visual proteins serve as models for human retinal diseases, and in environmental risk assessment where the impact of pollutants on marine visual function is increasingly recognized [6]. This application note provides a comprehensive framework for studying marine visual anatomy, with detailed protocols for microspectrophotometry, morphological analysis, and environmental assessment.
Photoreceptor Diversity and Visual Pigments
Photoreceptor Types and Distribution
Marine species exhibit extraordinary diversity in photoreceptor types and organization, reflecting adaptations to their specific ecological niches and visual requirements.
Table 1: Photoreceptor Types in Marine Species
| Species | Photoreceptor Types | Distribution Pattern | Ecological Adaptation |
|---|---|---|---|
| Atlantic Halibut (Hippoglossus hippoglossus) | Single, double, and triple cones | Square mosaic with regional variations | Demersal life style post-metamorphosis [2] |
| Molly Fish (Poecilia sphenops) | Double cones with two single cone variants | Square mosaic pattern throughout retina | Diurnal activity; motion awareness [7] |
| Sea Urchin (Echinoidea) | Distributed photoreceptors with opsins | All-body photoreceptor network | Whole-body light sensitivity without eyes [8] |
| Marine Protists | Four newly discovered photoreceptor groups | Cellular distribution | Daylight synchronization for growth cycles [9] |
In teleost fish like the Atlantic halibut, the retina undergoes dramatic reorganization during metamorphosis from larval to juvenile stages. Pre-metamorphic larvae typically possess a honeycomb mosaic of single cones, which transforms into a square mosaic of single and double cones in juveniles [2]. This transition enables adaptation from a pelagic to demersal lifestyle, with changes in diet from pelagic plankton to benthic invertebrates. The square mosaic configuration typically consists of four double cones arranged around a central single cone, optimizing both spatial resolution and chromatic sensitivity [7].
Visual Pigment Diversity
Visual pigments, consisting of opsin proteins bound to chromophores, determine the spectral sensitivity of photoreceptors. Microspectrophotometry has revealed remarkable diversity in marine visual pigments:
Table 2: Visual Pigments in Atlantic Halibut Identified by Microspectrophotometry
| Photoreceptor Type | Visual Pigment Class | Absorbance Peak (λmax, nm) | Probable Opsin |
|---|---|---|---|
| Short wavelength cones | S(431) | 431 nm | SWS1 or SWS2 |
| Short wavelength cones | S(457) | 457 nm | SWS2 |
| Middle wavelength cones | M(500) | 500 nm | RH2 |
| Middle wavelength cones | M(514) | 514 nm | RH2 |
| Middle wavelength cones | M(527) | 527 nm | RH2 |
| Long wavelength cones | L(550) | 550 nm | LWS |
| Rods | Rod pigment | 491 nm | RH1 [2] |
Sea urchins exemplify an extreme adaptation, where the entire body functions as a diffuse photoreceptive structure. They express multiple opsin types and neurotransmitters throughout their nervous system, creating what researchers term an "all-body brain" for distributed light processing [8]. This system enables urchins to respond to light despite lacking discrete eyes, with photoreceptors distributed across their body surface.
Experimental Protocols
Microspectrophotometry for Visual Pigment Analysis
Principle: Microspectrophotometry enables non-destructive measurement of absorbance spectra from individual photoreceptor cells, providing direct data on visual pigment spectral sensitivity.
Reagents and Equipment
- Marine species of interest (freshly collected or properly preserved)
- Physiological saline appropriate for the species
- Glutaraldehyde or other mild fixatives (optional, for stabilization)
- Liquid nitrogen for rapid freezing (if required)
- Multichannel microspectrophotometer system [4] [5]
- Microscope with infrared illumination and imaging capability
- Vibration isolation table to minimize spectral artifacts [4]
- Monochromator or other tunable light source (wavelength range 300-700 nm)
- Computer system with spectral acquisition and analysis software
Procedure
Sample Preparation
- Dark-adapt specimens for at least 2 hours prior to dissection
- Dissect retinal tissue under dim red light using microsurgical tools
- Prepare photoreceptor suspension by gentle mechanical agitation or enzymatic treatment (e.g., papain or hyaluronidase)
- Transfer suspension to microscope slide with coverslip sealed with petroleum jelly to prevent dehydration
Instrument Calibration
- Perform wavelength calibration using standard absorbance filters or rare-earth oxides
- Establish baseline with clear area of slide
- Verify system stability with control measurements
Spectral Measurements
- Identify target photoreceptors using infrared viewing system
- Align measuring beam (typically 1-2 μm diameter) with outer segment
- Scan through wavelength range (e.g., 700-300 nm) to record absorbance spectrum
- Expose to bright light to bleach photopigment and repeat scan
- Calculate difference spectrum to identify visual pigment absorbance [4]
Data Analysis
Retinal Histology and Photoreceptor Mosaic Analysis
Principle: Detailed morphological analysis of retinal structure reveals the organization of photoreceptor mosaics and their relationship to visual function.
Reagents and Equipment
- Fixative solutions (e.g., Bouin's fluid, paraformaldehyde-glutaraldehyde combination) [7]
- Ethanol series for dehydration (50%, 70%, 90%, 100%)
- Embedding media (paraffin for light microscopy, Araldite for electron microscopy)
- Staining solutions (toluidine blue, hematoxylin and eosin, lead citrate, uranyl acetate)
- Microtomes (rotary for paraffin, ultramicrotome for thin sections)
- Light and electron microscopes with digital imaging capabilities
Procedure
Tissue Fixation and Processing
- Dissect eyes and immerse in appropriate fixative within minutes of sacrifice
- For Bouin's fixation: 22 hours at room temperature [7]
- For ultrastructural analysis: 2.5% paraformaldehyde-glutaraldehyde combination for 24 hours [7]
- Dehydrate through graded ethanol series, clear with xylene or propylene oxide
- Infiltrate and embed in appropriate medium
Sectioning and Staining
- Cut serial sections at 4-5 μm thickness for light microscopy [7]
- Cut ultrathin sections (70 nm) for transmission electron microscopy [7]
- Stain paraffin sections with Harris hematoxylin and eosin for general morphology
- Stain semithin sections with toluidine blue for photoreceptor identification
- Stain ultrathin sections with uranyl acetate and lead citrate for TEM [7]
Mosaic Analysis
- Identify retinal regions (dorsal, ventral, nasal, temporal)
- Map photoreceptor distribution and density using image analysis software (e.g., ImageJ)
- Classify mosaic patterns (honeycomb, square, row) based on geometric arrangement
- Correlate mosaic organization with spectral data from MSP
Oil Droplet Adhesion Assessment for Ecological Risk
Principle: Quantifying oil droplet adhesion to marine organisms, particularly fish eggs, provides critical data for environmental risk assessment of oil spills.
Reagents and Equipment
- Fish eggs with adhesive chorion (e.g., haddock, cod)
- Crude oil samples or prepared oil-seawater dispersions
- Dispersant chemicals (e.g., Corexit) for comparison studies
- Flow-through exposure systems with precise temperature control
- Microscopy systems for documenting droplet adhesion
- Chemical analysis equipment for hydrocarbon quantification
Procedure
Experimental Setup
- Prepare oil-seawater dispersions with controlled droplet size distributions
- Characterize droplet size using Coulter counter or laser diffraction
- Measure zeta potential of oil droplets [10]
- Establish flow-through exposure system simulating natural conditions
Exposure Experiments
- Expose adhesive fish eggs to oil dispersions of known concentration
- Vary exposure duration, oil concentration, and droplet size systematically
- Include controls with dispersants alone and untreated seawater
- Maintain appropriate temperature and water quality throughout exposure
Adhesion Quantification
- Document droplet adhesion using microscopy with digital imaging
- Quantify accumulated oil mass per egg using chemical extraction and GC-MS
- Measure developmental abnormalities and mortality rates
- Calculate adhesion probabilities and exposure thresholds [6]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Marine Vision Studies
| Reagent/Category | Specific Examples | Research Application | Function in Protocol |
|---|---|---|---|
| Fixatives | Bouin's fluid, Paraformaldehyde-glutaraldehyde, Hartman's fixative [11] [7] | Tissue preservation for histology | Maintain structural integrity during processing |
| Embedding Media | Paraffin, Araldite resin [7] | Sectioning support | Provide structural support for thin sectioning |
| Staining Solutions | Toluidine blue, Hematoxylin & Eosin, Uranyl acetate, Lead citrate [7] | Tissue contrast enhancement | Highlight cellular structures for microscopy |
| Molecular Probes | Anti-GFAP, Anti-Rhodopsin, Anti-Calbindin, ViewRNA probes [11] [7] | Cellular localization | Identify specific proteins or mRNA in tissue |
| Enzymes | Papain, Hyaluronidase | Photoreceptor isolation | Dissociate retinal tissue for single-cell analysis |
| Buffers | Phosphate buffer, Sodium citrate, Tris-EDTA [11] | pH and osmotic maintenance | Maintain physiological conditions during processing |
| Optical Standards | Rare-earth oxides, Absorbance filters | MSP calibration | Verify wavelength accuracy and system performance |
Advanced Techniques and Applications
Molecular Histology Integration
Combining microspectrophotometry with molecular techniques provides a comprehensive understanding of visual system function:
Single-molecule Fluorescence In Situ Hybridization (smFISH) enables precise localization of opsin mRNA transcripts within retinal sections. When combined with immunofluorescence for opsin proteins and MSP for functional characterization, this approach reveals relationships between gene expression, protein localization, and spectral sensitivity [11]. Critical steps include:
- RNAse-free handling throughout sample processing
- Antigen retrieval pretreatments for formalin-fixed tissues
- Multiplexed detection with spectrally distinct fluorophores
- Controls for specificity and autofluorescence
Environmental Impact Assessment
The adhesion of oil droplets to marine organisms represents a significant environmental threat that requires specialized assessment protocols [6]:
Field Sampling
- Collect water samples during and after oil spills
- Document oil droplet size distributions and concentrations
- Capture affected organisms for adhesion assessment
Laboratory Analysis
- Quantify hydrocarbon composition of adhered droplets
- Measure developmental impacts on early life stages
- Determine threshold concentrations for adverse effects
Modeling Exposure Scenarios
- Implement individual-based models of organism movement
- Simulate oil dispersion under various environmental conditions
- Predict population-level impacts from individual exposure data
The integrated study of photoreceptors, retinal mosaics, and environmental interactions provides crucial insights into marine visual systems. Microspectrophotometry serves as a cornerstone technique, enabling direct measurement of visual pigment function that can be correlated with molecular, anatomical, and ecological data. The protocols outlined in this application note provide a comprehensive framework for investigating marine vision across multiple levels of biological organization, from molecular interactions to ecosystem-scale environmental impacts. As research in this field advances, these methodologies will continue to refine our understanding of how marine organisms perceive their visual world and respond to environmental challenges.
Spectral tuning is a fundamental evolutionary process whereby the visual systems of marine species adapt to the specific light qualities of their ecological niches. In aquatic environments, water acts as a spectral filter, selectively absorbing and scattering wavelengths, which creates diverse light habitats from surface waters to the deep sea [12]. This application note details the mechanisms of spectral tuning and provides standardized protocols for investigating visual adaptations in marine species, with a specific focus on microspectrophotometry (MSP) as a core analytical technique. The information is framed within the context of a broader thesis research project utilizing MSP for visual pigment analysis, providing essential methodologies and reagent solutions for researchers and scientists in the field.
Key Mechanisms of Spectral Tuning
Evolution has equipped marine organisms with multiple, often complementary, mechanisms to fine-tune their visual sensitivity to ambient light. The table below summarizes the primary spectral tuning mechanisms identified in marine fish and other vertebrates.
Table 1: Key Mechanisms of Spectral Tuning in Marine Species
| Mechanism | Functional Principle | Biological Example | Spectral Effect |
|---|---|---|---|
| Amino Acid Substitutions in Opsins | Changes in key amino acids in the opsin protein alter the interaction with the chromophore [12]. | Convergent evolution in primate LWS opsins and Characiform fish LWS-paralogs [12]. | Large (e.g., ~75 nm) or small (2-10 nm) shifts in λmax [12]. |
| A1/A2-Chromophore Shift | Switching the chromophore from 11-cis retinal (A1) to 11-cis 3,4-dehydroretinal (A2) [13]. | Marine fish like the masked greenling (Hexagrammos octogrammus) and prickleback (Pholidapus dybowskii) [13]. | Red-shifts the λmax of the visual pigment [12] [13]. |
| Gene Duplication & Loss | Whole-genome or gene-specific duplications provide genetic material for neofunctionalization [12]. | Characiform-specific duplication of LWS- and RH1-opsins following teleost-specific genome duplication (TGD) [12]. | Creates new visual pigments with distinct spectral sensitivities [12]. |
| Differential Opsin Expression | Varying the relative expression levels of different opsin genes in the retina [12]. | Characiforms base color vision on expression of LWS-paralogs and SWS2 [12]. | Modifies the retina's overall spectral sensitivity without changing pigment λmax. |
| Optical Filters (Oil Droplets) | Colored oil droplets in cone photoreceptors act as long-pass filters [14]. | Avian species like the whooping crane; R-type droplets (λcut 576 nm) in LWS cones [14]. | Narrows photoreceptor spectral sensitivity and reduces overlap between cone types [14]. |
Experimental Protocols for Visual Pigment Analysis
Protocol: Microspectrophotometry (MSP) of Photoreceptors
Objective: To measure the absorbance spectrum (λmax) of visual pigments in individual retinal photoreceptor cells.
Materials:
- Dark-adapted retinal tissue
- Physiological saline solution
- Microspectrophotometer
- Glass slides and cover slips
- Software for template fitting (e.g., Govardovskii et al. 2000 templates)
Procedure:
- Tissue Preparation: Euthanize the specimen following approved IACUC protocols. Enucleate the eye and hemisect it under dim red light. Remove the retina and place it in physiological saline.
- Cell Suspension: Gently homogenize the retinal tissue to create a suspension of individual photoreceptor cells.
- MSP Measurement: Transfer a small aliquot to a microscope slide. Using the microspectrophotometer, target single rods or cones and measure the absorbance spectrum across visible wavelengths.
- Data Analysis: Fit the recorded absorbance spectrum to known visual pigment templates to determine the λmax. For pigments containing an A1/A2 mixture, the data may be best fitted by a combination of templates [13].
Protocol: High-Performance Liquid Chromatography (HPLC) for Chromophore Analysis
Objective: To quantify the relative proportions of A1 (11-cis retinal) and A2 (11-cis 3,4-dehydroretinal) chromophores in the retina.
Materials:
- Retinal tissue
- HPLC system with a UV-Vis detector
- Normal-phase HPLC column
- Organic solvents (e.g., hexane, ethyl acetate)
- Standard solutions of A1 and A2 chromophores
Procedure:
- Chromophore Extraction: Homogenize the retinal tissue in a buffer. Extract retinoids using organic solvents.
- Derivatization: Convert the chromophores to their corresponding oximes to stabilize the 11-cis isomers.
- HPLC Analysis: Inject the sample onto the HPLC column. Elute using a gradient of organic solvents and detect the oximes at 360 nm.
- Quantification: Identify A1 and A2 oxime peaks by comparing their retention times to known standards. Calculate the A1:A2 ratio based on peak areas [13].
Protocol: Investigating Chromophore Conversion with Light/Dark Adaptation
Objective: To study the effects of light regime on the A1/A2 chromophore ratio in marine fish retinas.
Materials:
- Live fish specimens
- Controlled environment aquaria with adjustable lighting
- Equipment for MSP and HPLC (as above)
Procedure:
- Acclimation: Maintain one group of fish under a bright light adaptation regime (e.g., 12h light/12h dark) and another under dark adaptation (e.g., continuous darkness) for a period of 4-5 weeks. Control for water temperature.
- Sample Collection: After the acclimation period, euthanize the fish and harvest retinal tissues under dim red light.
- Analysis: Perform MSP and HPLC analysis on the retinal samples as described in Protocols 3.1 and 3.2.
- Comparison: Compare the λmax values from MSP and the A1:A2 ratios from HPLC between the light- and dark-adapted groups. Note: Some marine fish show an unusual increase in A2 proportion after light exposure, a reversal of the common pattern [13].
The Scientist's Toolkit: Research Reagent Solutions
The following table lists essential materials and reagents for conducting research on spectral tuning and visual pigment analysis.
Table 2: Essential Research Reagents and Materials for Visual Pigment Analysis
| Item | Function/Application | Example/Notes |
|---|---|---|
| 11-cis Retinal (A1) | Chromophore standard for HPLC; reconstitution of visual pigments in vitro [12] [13]. | Essential for determining A1:A2 ratio and for in vitro expression studies. |
| 11-cis 3,4-Dehydroretinal (A2) | Chromophore standard for HPLC; reconstitution of visual pigments in vitro [13]. | Used to quantify the A2 component in retinal extracts. |
| Opsin Expression Vectors | In vitro synthesis of visual pigments for spectral characterization [14]. | Allows for the measurement of λmax without the need for native tissue. |
| Govardovskii Template Equations | Computational tool for determining λmax from MSP absorbance data [13]. | Standard method for analyzing MSP spectral data. |
| Physiological Saline Solution | Maintenance of retinal tissue viability during dissection and preparation. | Typically an isotonic, buffered solution. |
| Normal-Phase HPLC Column | Chromatographic separation of A1 and A2 chromophore oximes [13]. | Critical for accurate quantification of chromophore ratios. |
Experimental Workflow and Chromophore Pathway
The following diagram illustrates the logical workflow for a comprehensive research project investigating spectral tuning, integrating the protocols described above.
Research Workflow for Spectral Tuning Analysis
The biochemical pathway of chromophore conversion is central to one key mechanism of spectral tuning. The following diagram outlines this process.
A1/A2 Chromophore Conversion Pathway
Microspectrophotometry (MSP) is an indispensable technique in marine visual ecology, enabling researchers to characterize visual pigments in aquatic species by measuring their spectral absorbance properties. This methodology provides critical insights into how marine organisms perceive their light-attenuated underwater environments. The technique's particular value lies in its ability to analyze minute retinal samples directly, yielding precise data on photoreceptor spectral sensitivity that correlates with specific visual behaviors and ecological adaptations. For marine researchers investigating visual pigment diversity, MSP offers the resolution needed to document the complex visual systems that have evolved in response to the unique photic conditions of aquatic habitats.
The integration of multichannel detectors with MSP systems has significantly advanced the field, allowing for simultaneous measurements across multiple wavelengths and dramatically improving data acquisition efficiency. This technological synergy is especially valuable when working with marine species that may possess multiple visual pigment classes within a single retina—a common adaptation to variable light conditions at different depths. This application note details the essential toolkit and methodologies for employing MSP in marine visual pigment research, providing structured protocols, data presentation standards, and analytical frameworks tailored to the unique challenges of aquatic visual systems.
Technical Specifications & Data Presentation
Key Performance Metrics for Marine Visual Pigment Analysis
Microspectrophotometers deployed in marine visual research must meet specific performance criteria to accurately characterize the diverse visual pigments found in aquatic species. The following table summarizes essential technical specifications optimized for visual pigment analysis:
Table 1: Microspectrophotometer Technical Specifications for Visual Pigment Analysis
| Parameter | Specification | Application Notes |
|---|---|---|
| Wavelength Range | 350-750 nm | Covers UV to far-red spectrum; essential for marine species with UV sensitivity [15] |
| Spectral Bandwidth | ≤ 2 nm | Sufficient resolution to distinguish closely-spaced visual pigment λmax values |
| Photometric Accuracy | ±0.003 absorbance units | Critical for detecting small absorbance changes in visual pigment measurements |
| Spatial Resolution | 1-2 μm spot size | Enables measurement of individual photoreceptor outer segments |
| Detector Type | Multichannel CCD or CMOS | Simultaneous multi-wavelength detection reduces measurement time and photobleaching [15] |
| Beam Switching Frequency | 200-400 Hz | Minimizes photopigment bleaching during scanning procedures |
Quantitative Visual Pigment Data from Marine Species
MSP analysis of marine species reveals remarkable diversity in visual pigment composition, reflecting adaptations to specific photic environments. The following table presents representative absorbance data (λmax values) from various marine taxa:
Table 2: Visual Pigment Absorbance Maxima (λmax) in Marine Species
| Species | Photoreceptor Type | Visual Pigment λmax (nm) | Ecological Context |
|---|---|---|---|
| Atlantic Halibut (Hippoglossus hippoglossus) | Rod | 491 | Demersal lifestyle; metamorphic visual system reorganization [2] |
| SWS Cone | 431, 457 | Multiple short-wave pigments for enhanced contrast in blue-shifted marine light | |
| MWS Cone | 500, 514, 527 | Middle-wavelength discrimination in variable depth habitats | |
| LWS Cone | 550 | Potential benthic prey detection against substrate | |
| Winter Flounder (Pseudopleuronectes americanus) | Pre-metamorphic Cone | 519 | Larval pelagic phase visual requirements [2] |
| Post-metamorphic SWS Cone | 457 | Benthic juvenile phase with expanded spectral sensitivity | |
| Post-metamorphic LWS Cone | 531, 547 | Substrate discrimination and predator detection |
Experimental Protocols
Sample Preparation Protocol for Marine Retinal Tissue
Principle: Proper preparation of retinal tissue from marine species is critical for preserving visual pigment integrity and obtaining accurate spectral measurements. This protocol outlines standardized procedures for handling diverse marine retinal samples.
Materials:
- Marine Ringer's solution (species-appropriate osmolarity)
- Phosphate-buffered saline (PBS, 0.1 M, pH 7.4)
- Glutaraldehyde solution (2.5% in buffer)
- Microscope slides and coverslips
- Tungsten or glass microtools for dissection
- Liquid nitrogen for flash freezing (optional)
Procedure:
Tissue Extraction and Initial Processing
- Euthanize specimen following institutional animal care guidelines
- Enucleate eyes and hemisect at the ora serrata under dim red light
- Remove vitreous humor carefully using fine forceps
- Isolate retinal tissue and transfer to marine Ringer's solution
Photoreceptor Isolation
- For rod-dominant species: gently agitate retinal pieces to dissociate outer segments
- For cone-dominant species: use fine vibration or enzymatic digestion (0.01% trypsin for 5-10 minutes) to liberate photoreceptors
- For flatfish species during metamorphosis: note retinal quadrant origin due to mosaic heterogeneity [2]
Sample Mounting
- Transfer suspension to clean microscope slide
- Allow photoreceptors to settle for 2-3 minutes
- Carefully aspirate excess fluid without allowing complete drying
- Apply coverslip, sealing edges with vacuum grease to prevent dehydration
Quality Control Checks
- Verify photoreceptor structural integrity using differential interference contrast microscopy
- Assess for bleaching by comparing initial and post-measurement absorbance
- Document sample orientation and photoreceptor type (rod, single cone, double cone)
Troubleshooting Notes:
- If photoreceptors show signs of bleaching, implement more stringent dim-light conditions
- For samples with poor adhesion, use poly-L-lysine coated slides
- When measuring multiple cone types, document spatial relationships for mosaic analysis
MSP Measurement Protocol for Visual Pigment Characterization
Principle: This protocol details the specific steps for acquiring absorbance spectra from marine photoreceptors using microspectrophotometry, with emphasis on preserving pigment integrity and ensuring measurement accuracy.
Materials:
- Microspectrophotometer with multichannel detector
- Calibration standards (didymium glass, neutral density filters)
- Data acquisition software with bleaching correction algorithms
- Reference beam neutral density filter (if required by system)
Procedure:
System Calibration
- Wavelength calibration using holmium oxide or didymium glass standard
- Photometric calibration using neutral density filters of known transmission
- Background measurement with clear area of slide adjacent to sample
Sample Alignment and Measurement
- Locate intact photoreceptor using dim infrared illumination with image converter
- Align measuring beam to pass through outer segment longitudinally
- Position reference beam through clear area adjacent to cell
- For double cones in marine species: align beam through individual members separately [2]
Spectral Scanning
- Acquire baseline spectrum from 750nm to 350nm (reverse scan reduces bleaching)
- Repeat scan to verify stability (difference should be <0.005 absorbance units)
- Expose sample to bright white light for 3-5 minutes to bleach visual pigment
- Acquire post-bleach spectrum using identical parameters
Data Integrity Verification
- Calculate difference spectrum (pre-bleach minus post-bleach)
- Verify spectrum meets template fit criteria for visual pigments
- Discard data showing evidence of photoproduct accumulation or photobleaching during initial scan
Technical Notes:
- For species with multiple visual pigments, ensure measuring beam is properly aligned to target specific photoreceptor types
- When working with very small cones from marine fish, reduce beam size to minimize contamination from adjacent cells
- For accurate λmax determination, maintain signal-to-noise ratio >10:1 at peak absorbance
Data Analysis Framework
Functional Data Analysis for Spectral Interpretation
Principle: Functional data analysis provides a robust statistical framework for evaluating MSP evidence, treating absorbance spectra as continuous functions rather than discrete measurements. This approach enhances quantitative comparisons between marine visual pigments.
Likelihood Ratio Calculation: The evidence for whether control and recovered spectra originate from the same source is evaluated using the likelihood ratio (LR), computed as follows [15]:
Where:
- zc and zr represent transformed control and recovered spectral data
- θ represents the group mean with normal distribution N(η, C)
- U represents the fixed covariance matrix for within-group distribution
Implementation Steps:
Spectral Data Transformation
- Represent absorbance spectra using B-spline basis functions: f(x) ≈ Σθₖφₖ(x)
- Select optimal number of basis functions (B) and polynomial order (o) using Akaike's Information Criterion [15]
- Standardize wavelength axis to [0,1] interval to facilitate comparisons
Multivariate Random Effects Modeling
- Model within-group distribution as Z ∼ N(θ, U)
- Model between-group distribution as θ ∼ N(η, C)
- Estimate fixed covariance matrices U and C from training data
Hypothesis Testing Framework
- Hp: Control and recovered spectra originate from same source
- Hd: Control and recovered spectra originate from different sources
- Compute LR support for propositions using multivariate normal density functions
Research Reagent Solutions
Table 3: Essential Research Reagents for Marine Visual Pigment Analysis
| Reagent/Material | Specification | Application Function |
|---|---|---|
| Marine Ringer's Solution | Species-specific osmolarity (900-1100 mOsm) | Maintain physiological ionic environment during tissue preparation |
| Digestive Enzymes | Trypsin (0.01-0.1%), Collagenase (0.05%) | Liberate photoreceptors from retinal tissue for individual analysis |
| Fixatives | Glutaraldehyde (2.5% in buffer), Paraformaldehyde (4%) | Structural stabilization for morphological correlation (limited use for MSP) |
| Mounting Media | Glycerol in PBS, Polyvinyl alcohol | Non-fluorescent media for sample preservation during measurement |
| Neutral Density Filters | Certified transmission values (0.1-3.0 OD) | Photometric calibration and reference beam attenuation |
| Wavelength Standards | Holmium oxide, Didymium glass | Accurate wavelength calibration across UV-visible spectrum |
| Opsin Antibodies | Species-specific where available | Immunohistochemical validation of opsin expression patterns |
Workflow Visualization
Marine Visual Pigment Analysis Workflow
Data Processing Pathway
The integrated toolkit of microspectrophotometry with multichannel detection provides marine researchers with a powerful methodology for elucidating visual pigment diversity in aquatic species. The protocols and analytical frameworks presented here offer standardized approaches for collecting robust spectral data while accounting for the unique challenges of marine visual systems. The application of functional data analysis represents a significant advancement over traditional qualitative assessments, enabling quantitative evaluation of spectral evidence within likelihood ratio frameworks. As marine visual ecology continues to explore the complex relationships between visual pigment complement, habitat depth, light environment, and behavioral ecology, these refined MSP methodologies will be essential for documenting and interpreting the remarkable visual adaptations that enable marine species to thrive in diverse photic environments.
From Ship to Lab: MSP Methodologies for Marine Species Analysis
Microspectrophotometry (MSP) serves as a cornerstone technique in visual neuroscience, enabling the direct measurement of visual pigment absorbance within individual photoreceptor cells. This application note provides a detailed protocol for preparing delicate marine biological samples—specifically retinas and isolated rhabdoms—for MSP analysis. Proper sample preparation is paramount for obtaining accurate and reproducible spectral data, as the integrity of visual pigments is easily compromised by improper handling. The methodologies outlined herein are framed within a broader thesis on advancing MSP for marine visual ecology research, offering researchers a standardized approach for investigating the fascinating diversity of marine visual systems.
The primary objective of sample preparation for MSP is to preserve the native state of visual pigments and the structural integrity of photoreceptors until spectral measurements can be performed. Marine samples present unique challenges:
- Rapid Degradation: Visual pigments are inherently photosensitive and begin degrading immediately upon exposure to light post-mortem. This is exacerbated in marine species, whose pigments may be adapted to specific, often cold, environmental conditions [13].
- Structural Fragility: The retinal tissue and rhabdoms of many marine invertebrates and fish are exceptionally soft and easily damaged during dissection and manipulation [16].
- Spectral Complexity: Many marine organisms possess mixed visual pigment systems (e.g., A1/A2 chromophore mixtures) that can be labile and alter with light exposure or temperature changes, requiring preparation techniques that stabilize the native pigment ratio [13].
Success hinges on three core principles: working under appropriate lighting conditions, maintaining physiological conditions where possible, and minimizing mechanical stress on the sample.
Essential Reagents and Equipment
The following toolkit is essential for the preparation of marine retinal samples. Specific reagents mentioned in the literature are summarized in the table below.
Table 1: Research Reagent Solutions for Marine Retina Preparation
| Reagent/Material | Function | Example from Literature |
|---|---|---|
| Glutaraldehyde | Light chemical fixation of tissue sections; promotes photobleaching and stabilizes rhabdom structure. | Used at 0.5% in filtered seawater for stomatopod larval sections [16]. |
| Filtered Seawater | Physiological mounting and dissection medium for marine tissue. | Used as a solvent for the glutaraldehyde fixative [16]. |
| Silicone Grease | Creating a sealed chamber between coverslips to contain the sample and mounting medium. | Used to mount cryosections for MSP measurement [16]. |
| Cryostat (Cryomicrotome) | Sectioning frozen retinal tissue into thin slices for MSP measurement. | Used at -30°C to section flash-frozen stomatopod eyes [16]. |
| Difluoroethane Spray | Rapid freezing of dissected tissue to preserve its state and prevent ice crystal formation. | Used to flash-freeze dissected eyes prior to cryosectioning [16]. |
Step-by-Step Experimental Protocol
Animal Handling and Dark Adaptation
- Dark Adaptation: Prior to sacrifice, animals must be fully dark-adapted to ensure visual pigments are in their native, unbleached state. Place the animal in a light-tight container for a period appropriate to its biology. For example, late-stage stomatopod larvae were dark-adapted for a minimum of 3 hours, while early-stage larvae were adapted overnight [16].
- Sacrifice: All subsequent steps must be performed under dim red light, to which most marine visual pigments are insensitive. Sacrifice the animal according to approved ethical guidelines.
Dissection and Tissue Isolation
- Rapid Dissection: Under dim red light illumination, quickly dissect the eyes from the animal. Speed is critical to prevent metabolic changes and post-mortem degradation.
- Primary Fixation/Freezing: Immediately process the isolated eyes. For cryosectioning, flash-freeze the tissue using a cryogen such as difluoroethane spray [16]. Alternatively, some protocols may use light chemical fixation.
Preparation of Retinal Sections
- Cryosectioning: Mount the flash-frozen tissue in a cryomicrotome maintained at -30°C. Section the tissue at a thickness of 10–12 µm [16]. This thickness is optimal for MSP, providing enough pigment density for measurement without excessive light scattering.
- Section Mounting: Transfer the sections to a microscope slide and lightly fix them in a solution of 0.5% glutaraldehyde in filtered seawater for approximately 10 minutes [16]. This step helps stabilize the tissue structure and promotes controlled photobleaching during MSP analysis.
- Coverslip Sealing: Mount the fixed section between two coverslips using a ring of silicone grease to create a sealed chamber containing the mounting medium. This prevents the sample from drying out during measurement.
Preparation of Isolated Rhabdoms
While the search results provide more detail on retinal sections, the preparation of isolated rhabdoms for MSP, as referenced in historical studies, generally follows this workflow [17]:
- Retina Isolation: Dissect the retina under dim red light and place it in a suitable physiological saline.
- Mechanical Separation: Gently homogenize or shred the retinal tissue to release the rhabdoms from the photoreceptor cells.
- Chemical Fixation: Immerse the isolated rhabdoms in a low-concentration glutaraldehyde solution (e.g., as used in fixed drone retinae [17]) to stabilize their structure.
- Mounting: Transfer a small aliquot of the suspension containing rhabdoms onto a microscope slide for MSP measurement.
The following workflow diagram synthesizes the core sample preparation pathway based on the protocols described above.
Data Presentation and Analysis
The ultimate goal of sample preparation is to yield high-quality data on visual pigment absorbance. MSP measurements generate spectral curves, but the preparation method itself is defined by specific parameters. The table below quantifies key aspects of the preparation protocols as found in the literature.
Table 2: Quantitative Parameters for Sample Preparation from Literature
| Protocol Step | Key Parameter | Reported Value / Specification | Context |
|---|---|---|---|
| Dark Adaptation | Duration | Minimum 3 hours to overnight [16] | For stomatopod larvae |
| Cryosectioning | Section Thickness | 10–12 µm [16] | Standard for MSP analysis |
| Cryosectioning | Chamber Temperature | -30°C [16] | For stomatopod larval eyes |
| Chemical Fixation | Glutaraldehyde Concentration | 0.5% [16] | In filtered seawater |
Troubleshooting and Technical Notes
- Poor Signal-to-Noise in MSP: This can result from over-fixation, which destroys the visual pigment, or under-fixation, leading to poor structural integrity. The 0.5% glutaraldehyde concentration is a critical starting point [16].
- Sample Degradation: If pigments appear bleached upon measurement, check for light leaks during dissection or insufficient dark adaptation time. Ensure all steps are performed under strict dim red light conditions.
- Tissue Damage During Sectioning: If the retina is tearing, the freezing process may be too slow, leading to large ice crystal formation. Ensure the tissue is flash-frozen as rapidly as possible after dissection.
Ship-based microspectrophotometry (MSP) is a powerful technique for the in-situ characterization of visual pigments in marine species, enabling researchers to obtain crucial data on the spectral absorbance properties of photoreceptors directly in the field. This methodology is particularly valuable for studies on marine vertebrates and invertebrates where prompt measurement following capture is essential to preserve the native state of visual pigments [18] [19]. However, the marine environment introduces significant technical challenges, with vibration artifacts representing a primary obstacle to obtaining reliable spectral data. Shipboard vibrations, originating from propulsion systems, generators, wave impacts, and onboard machinery, can severely compromise the precision of optical measurements by introducing misalignment, noise, and instability in the microspectrophotometer [20] [21]. These issues are particularly pronounced for modern vessel designs, which have become more optimized and consequently more susceptible to vibration-induced failures that can affect sensitive instrumentation [21].
The problem of structural vibration in ships is well-documented, with flat plate structures constituting the basic structural units of hull components such as cabins, double bottoms, and superstructures [20]. These vibrations propagate through the ship's structure and can disrupt the critical alignment required for measuring minute absorbance spectra in photoreceptor cells. For MSP applications, where measurements often target single cone cells or rods with diameters of just a few micrometers, even sub-micron vibrations can render data unusable [18] [2]. This application note provides a comprehensive framework for mitigating vibration artifacts in ship-based MSP systems, ensuring the collection of research-grade spectral data for visual pigment analysis in marine species.
Vibration Characteristics in Marine Environments
Understanding the origin and transmission pathways of shipboard vibrations is fundamental to developing effective mitigation strategies. Marine vessels present a complex vibration landscape with multiple excitation sources operating across different frequency ranges. The primary sources include rotating machinery such as motors, pumps, and fans; propulsion systems including shafts and propellers; and environmental forces from wave impacts and hydrodynamic flow [22] [21]. These vibrations propagate through the ship's structure, predominantly through flat plate components that form the basic structural elements of hull design [20]. The resulting vibrational energy distributes throughout the vessel, creating a challenging environment for sensitive optical instrumentation like microspectrophotometers.
Modern ship designs have exacerbated these challenges through structural optimizations that reduce weight and material usage while potentially increasing vibration susceptibility [21]. The problem is particularly acute in research vessels, which must accommodate both propulsion machinery and scientific equipment in a confined space. Vibration-induced failures in instrumentation can manifest as structural fatigue, component malfunction, and measurement inaccuracies - all of which directly impact the quality of scientific data collection [21]. For MSP systems, which depend on precise optical alignment and stability, these vibrations introduce noise, drift, and misalignment that compromise the integrity of visual pigment measurements.
Vibration Impact on Microspectrophotometry
The detrimental effects of vibration on MSP measurements are multifaceted and particularly problematic when working with the minute samples characteristic of marine organism photoreceptors. The wedge-tailed shearwater study, for instance, required measurements from seven different photoreceptor types including single cones with visual pigments ranging from violet (λmax 406 nm) to long-wavelength (λmax 566 nm) sensitivity [18] [23]. Achieving this resolution demands exceptional stability throughout the measurement process. Vibration-induced misalignment can cause beam deviation, leading to inaccurate absorbance measurements as the light path shifts relative to the microscopic sample. Low-frequency vibrations introduce baseline drift in spectral recordings, while higher-frequency components contribute signal noise that obscures the subtle spectral features of visual pigments.
The problem intensifies when measuring samples from deep-sea species, where visual pigments often exhibit spectral tuning to specific light environments. Studies on cetacean visual pigments have revealed precise spectral tuning of rod pigments to available light at foraging depths, with an inverse relationship observed between the wavelength of maximum sensitivity and depth [24]. Resolving these spectral characteristics requires vibration mitigation strategies that address the full frequency spectrum of shipboard vibrations. Furthermore, the trend toward rod monochromacy in multiple cetacean lineages (including sperm whales, some baleen whales, and beaked whales) means that researchers are often working with a single visual pigment type, making accurate characterization even more critical [25].
Table 1: Characteristic Vibration Sources and Their Impact on MSP Measurements
| Vibration Source | Frequency Range | Primary Impact on MSP | Affected Measurement Parameters |
|---|---|---|---|
| Main Engines & Propellers | 5-30 Hz | Low-frequency drift | Baseline stability, long-term measurements |
| Generators & Pumps | 30-100 Hz | Medium-frequency oscillation | Absorbance peak resolution, signal-to-noise ratio |
| Wave Impact & Hydrodynamic Forces | 1-20 Hz | Broad-spectrum instability | General measurement reliability, alignment |
| HVAC Systems | 10-50 Hz | Continuous vibration | Precision of λmax determination |
| Auxiliary Machinery | 50-200 Hz | High-frequency noise | Spectral fine structure, measurement precision |
Vibration Mitigation Framework
Instrument Isolation Protocols
Effective vibration mitigation begins with comprehensive instrument isolation designed to decouple the microspectrophotometer from the ship's structure. A multi-layered approach delivers the most significant improvement in measurement stability, particularly for the critical alignment components within the optical path. The primary isolation system should incorporate pneumatic isolators with low natural frequencies (0.5-5 Hz) to address the dominant low-frequency vibrations originating from propulsion systems and wave action [20]. These should be supplemented with damping materials such as sorbothane or specialized polymer composites at instrument contact points to absorb higher-frequency vibrations from generators and rotating machinery.
The optical bench within the MSP system requires specialized attention, as measurements of visual pigment absorbance in marine species demand exceptional stability. Research on marine ostracodes, for instance, identified visual pigment absorbance peaks at approximately 460 nm, with luminescence emission spectra peaking at 473 nm [19]. Resolving these spectral characteristics requires implementing internal passive isolation within the instrument itself, particularly for the monochromator, sample stage, and detector assemblies. Kinematic mounting of optical components, constrained layer damping on flat surfaces, and strategic use of damping alloys for component holders can significantly reduce vibration-induced misalignment and noise. For shipboard installations, additional inertial stabilization of the light source and reference beam path may be necessary, particularly when working with the high magnification objectives required for targeting individual photoreceptor cells.
Operational Procedures and Measurement Techniques
Adapting standard MSP protocols to compensate for residual vibration artifacts is essential for obtaining reliable data in shipboard environments. The measurement workflow should incorporate vibration-aware timing that synchronizes spectral scans with periods of relatively lower vibration, such as when the vessel is maintaining steady speed without course changes or when station-keeping in favorable sea states. During the larval to juvenile transition of Atlantic halibut, researchers documented complex photoreceptor reorganization involving the formation of square mosaics from single cones [2]. Studying such delicate morphological changes requires implementing signal averaging techniques with increased scan repetitions (typically 50-100% more than laboratory standards) to improve signal-to-noise ratio while applying adaptive filtering algorithms that recognize and reject vibration-corrupted scans in real-time.
Sample handling procedures must be optimized to minimize exposure to vibration during critical preparation and measurement phases. For marine species with diverse photoreceptor arrangements, such as the Atlantic halibut which exhibits six cone visual pigments with absorbance maxima ranging from 431 nm to 550 nm [2], precise positioning is essential. Implementing rapid mounting protocols that stabilize samples quickly and utilizing vibration-damped micropositioners for final alignment can preserve sample integrity while minimizing vibration exposure. Additionally, establishing a standardized vibration monitoring protocol using accelerometers mounted to the instrument frame provides quantitative data for correlating vibration levels with measurement quality, enabling post-processing compensation when necessary.
Table 2: Vibration Mitigation Solutions for Ship-Based MSP Components
| MSP Component | Primary Vulnerability | Recommended Solution | Performance Metric |
|---|---|---|---|
| Optical Bench | Structural resonance | Kinematic mounting, damping composites | Reduction of resonant amplification by >60% |
| Light Source | Filament/mirror stability | Secondary isolation platform | Intensity fluctuation <0.5% during scans |
| Monochromator | Grating alignment | Temperature compensation, stiffened mounts | Wavelength shift <0.1 nm during measurement |
| Sample Stage | Positioning accuracy | Piezo-electric stabilization, damped controllers | Positional drift <0.2 μm during single scan |
| Detection System | Detector noise | Vibration-resistant housing, EMI shielding | Noise reduction of 40-60% across spectrum |
| Data Acquisition | Timing errors | Vibration-triggered scan rejection | Automatic rejection of >90% of corrupted scans |
Experimental Protocol for Ship-Based Visual Pigment Analysis
Sample Preparation Under Shipboard Conditions
Preparation of retinal samples for MSP analysis under shipboard conditions demands specialized protocols that account for both the marine environment and vibration constraints. For marine birds like the wedge-tailed shearwater, which possesses five different types of vitamin A1-based visual pigment across seven photoreceptor types [18], immediate processing following collection is essential to preserve pigment integrity. The dissection should be conducted in a stabilized workstation with vibration-damped surfaces, utilizing temperature-controlled chambers to maintain samples at species-appropriate conditions throughout preparation. For most marine species, this ranges between 2-8°C to slow metabolic processes without causing thermal damage to photoreceptor structures.
The mounting procedure requires particular attention to vibration mitigation. Retinal samples should be oriented and secured using UV-polymerizing adhesives rather than mechanical clamping to minimize stress and vibration transmission. For species with complex photoreceptor distributions, such as flatfishes undergoing metamorphic transition from honeycomb to square cone mosaics [2], sectioning should be performed with vibration-stabilized microtomes to preserve structural integrity. The final sample orientation must facilitate rapid identification of target photoreceptors – a particular challenge for deep-diving marine mammals like cetaceans, which may possess only a single visual pigment type [25] [24]. Implementing pre-alignment protocols that establish reference coordinates during calm periods significantly reduces measurement time when vibration conditions temporarily improve.
Vibration-Resilient Measurement Workflow
The spectral measurement protocol must be specifically adapted to compensate for persistent low-frequency vessel vibrations while maintaining the precision required for accurate visual pigment characterization. The following workflow integrates multiple vibration mitigation strategies:
System Stabilization Phase: Allow a minimum of 30 minutes for instrument warm-up and stabilization, with continuous monitoring of vibration levels using integrated accelerometers. During this period, perform preliminary alignment using stable reference materials that simulate sample properties.
Baseline Acquisition: Collect reference spectra with the sample translated clear of the beam path, immediately preceding each sample measurement series. This approach minimizes the impact of low-frequency drift in baseline measurements, which is particularly important for species with violet-sensitive visual pigments like the wedge-tailed shearwater (λmax 406 nm) where signal intensity is naturally lower [18].
Adaptive Scanning Protocol: Implement a scanning strategy that dynamically adjusts integration times based on real-time vibration monitoring. For relatively stable periods (vibration amplitude <0.5 μm), utilize standard integration times of 0.5-1.0 seconds per wavelength step. During higher vibration intervals, switch to shorter integration times (0.1-0.2 seconds) with increased repetition and averaging.
Multi-Spectral Validation: For each measurement location, collect overlapping spectral ranges with varying scan speeds to identify and exclude vibration-induced artifacts. This approach is particularly valuable when characterizing complex visual systems such as those in Atlantic halibut, which exhibits multiple middle-wavelength-sensitive pigments (M(500), M(514), and M(527)) that require precise differentiation [2].
Post-Measurement Verification: Immediately following each spectral measurement, verify sample integrity and positioning through rapid imaging or confirmatory scans. For shipboard analysis of marine ostracodes, which display visual pigment absorbance maxima at approximately 460 nm [19], this verification ensures that vibration has not displaced the measurement area during scanning.
Data Analysis and Artifact Correction
Vibration Identification in Spectral Data
Recognizing and quantifying vibration-induced artifacts in MSP spectra is essential for ensuring data quality and making informed decisions about measurement validity. Vibration corruption typically manifests as increased high-frequency noise, baseline irregularities, and peak position instability – each requiring different identification approaches. For marine species with multiple visual pigment classes, such as the wedge-tailed shearwater with its VS, SWS, MWS, and LWS cone types [18], the impact of vibration varies with the spectral region being measured. Short-wavelength measurements (e.g., violet-sensitive λmax 406 nm) generally show greater susceptibility to vibration-induced noise due to lower photon flux and detector sensitivity in these regions.
The most reliable indicator of vibration contamination is spectral inconsistency between repeated measurements of the same photoreceptor. Implementing a quantitative consistency metric based on the normalized root mean square deviation (NRMSD) between successive scans provides an objective measure of vibration impact. For quality control thresholds, measurements exceeding 5% NRMSD should trigger investigation, while those exceeding 10% warrant rejection and repetition. Additional vibration indicators include abnormal peak broadening beyond the instrument's characteristic point spread function and wavelength-dependent noise patterns that correlate with known vibration frequencies in the vessel. Establishing these criteria is particularly important when studying species like cetaceans, where spectral tuning of visual pigments to depth represents a key adaptive trait [24].
Computational Compensation Methods
When vibration artifacts cannot be eliminated through physical mitigation alone, computational approaches provide a secondary defense for recovering usable spectral data. Adaptive filtering algorithms that utilize reference signals from accelerometers mounted on the instrument can effectively remove vibration-correlated noise components from spectral measurements. These approaches are particularly valuable for long-term monitoring studies where environmental conditions inevitably vary. For species with complex visual pigment complements, such as Atlantic halibut with six cone visual pigments [2], maintaining consistent measurement quality across multiple spectral classes requires frequency-dependent noise suppression tailored to each pigment's characteristic absorbance range.
Spectral reconstruction techniques offer an alternative approach for recovering accurate λmax values from vibration-compromised data. These methods leverage a priori knowledge of visual pigment template shapes [19] to fit partially corrupted spectra, with confidence metrics derived from the goodness of fit to established pigment models. For rod-dominated visual systems like those found in many cetaceans [25] [24], where the spectral tuning range may be relatively constrained, Bayesian estimation methods can incorporate ecological context (e.g., typical foraging depths) to improve reconstruction accuracy. Implementation of these computational compensation methods requires careful validation against laboratory standards and should be documented transparently when reporting research findings.
Research Reagent Solutions for Marine Visual Pigment Analysis
Table 3: Essential Research Reagents for Ship-Based Visual Pigment Analysis
| Reagent/Material | Function | Shipboard Adaptation | Application Example |
|---|---|---|---|
| Dimethyl sulfoxide (DMSO) with stabilizers | Visual pigment extraction and stabilization | Pre-aliquoted, temperature-stable formulations in sealed vials | Extraction of rod visual pigments from cetacean retinas [24] |
| Hydroxylamine hydrochloride | Chromophore bleaching control | Oxygen-impermeable packaging with moisture control | Testing retinoid-based pigment stability in deep-sea fish [26] |
| Phosphate-buffered saline (PBS) with antioxidants | Tissue preservation during dissection | Pre-mixed, sterile single-use aliquots | Maintenance of photoreceptor integrity in marine bird retinas [18] |
| Siliconizing reagents | Surface treatment for sample holders | Low-volatility formulations for controlled application | Preventing adhesion of fragile photoreceptors in ostracode studies [19] |
| UV-curable optical adhesives | Sample mounting and stabilization | Vibration-resistant formulations with delayed curing | Securing retinal sections from flatfish during metamorphosis [2] |
| Deuterium-tungsten hybrid light source | Broad-spectrum illumination for MSP | Ruggedized design with reinforced filaments | Continuous spectrum for visual pigment mapping in marine species [18] |
| Spectral calibration standards | Wavelength accuracy verification | Stable, non-degrading materials in shock-resistant mounting | Daily validation of MSP precision for whale pigment studies [25] |
Validation and Quality Assurance Protocol
Performance Metrics and Verification Standards
Establishing rigorous quality assurance protocols is essential for validating ship-based MSP data, particularly when vibration mitigation strategies may introduce their own artifacts. The validation framework should incorporate daily system verification using stable spectral standards with known absorbance characteristics across the relevant wavelength range (typically 350-650 nm for marine visual pigments). For each analytical session, baseline performance metrics should include wavelength accuracy (deviation < ±0.5 nm), spectral resolution (bandwidth < 5 nm), and absorbance precision (CV < 2% for repeated measurements) under simulated vessel vibration conditions.
The validation protocol must include biological reference materials that approximate the sample matrix being studied. For marine fish visual pigment analysis, standardized retinal preparations from readily available species with well-characterized visual pigments provide essential method verification. Studies of Baltic Sea fishes, for instance, have documented rod visual pigment spectral shifts between marine and limnic populations [26], providing potential reference systems for method validation. Additionally, implementing intermediate precision testing that evaluates performance across different sea states and vessel operations provides crucial data on the robustness of the vibration mitigation strategy. This approach is particularly valuable for research vessels that transition between transit operations and stationary sampling, as vibration profiles differ significantly between these modes.
Documentation and Reporting Standards
Comprehensive documentation of vibration conditions and mitigation effectiveness ensures the scientific validity of ship-based MSP data and enables meaningful comparison with laboratory studies. Each measurement series should include vibration metadata capturing quantitative vibration levels (frequency spectra and amplitude distributions) during spectral acquisition, specific mitigation strategies employed, and any computational corrections applied during data processing. This documentation is particularly important when reporting subtle spectral differences, such as the rod visual pigment shifts observed between marine and Baltic Sea populations of herring, flounder, and sand goby [26].
The reporting framework should explicitly acknowledge the limitations imposed by the shipboard environment while demonstrating the effectiveness of mitigation strategies through quality control metrics. For studies investigating evolutionary adaptations in visual pigments, such as the parallel spectral tuning observed in whale M/LWS pigments [25], the methodology section should include sufficient detail on vibration control to establish measurement reliability. Implementing these documentation standards across the research community will facilitate the development of improved vibration mitigation strategies and enhance the credibility of ship-based MSP as a technique for in-situ visual pigment analysis.
This application note details the use of microspectrophotometry (MSP) to investigate the dynamic reorganization of photoreceptors and their visual pigments during the metamorphosis of Atlantic halibut (Hippoglossus hippoglossus). Metamorphosis in flatfish involves a dramatic transition from bilaterally symmetrical, pelagic larvae to asymmetrical, demersal juveniles, accompanied by profound changes in the retinal architecture and visual pigment complement [2]. This case study aligns with a broader thesis on applying MSP to understand visual ecology in marine species, demonstrating how precise photopigment measurement can reveal adaptations to shifting environmental demands and lifestyles.
The retinal transformation in Atlantic halibut involves a complex transition from a larval honeycomb mosaic of single cones to a juvenile square mosaic incorporating single and double cones [2]. Concurrently, MSP revealed an expansion of the visual pigment repertoire from a pre-metamorphic single cone pigment to at least six distinct cone visual pigments in the juvenile, alongside the emergence of rod pigments for scotopic (dim-light) vision [2]. These findings highlight the utility of MSP in correlating morphological restructuring with functional chromatic diversity, providing a model for studying sensory system plasticity.
Key Quantitative Findings
Temporal Development of Eye Morphology
Table 1: Morphometric changes in the Atlantic halibut eye during metamorphosis. Data sourced from [2].
| Developmental Stage (Accumulated Temperature Units, ATU) | Approximate Days Post-Fertilization | Eye Long Axis (mm) | Eye Short Axis (mm) | Lens Diameter (mm) |
|---|---|---|---|---|
| 720 ATU | 96 days | 1.6 ± 0.18 | 1.3 ± 0.11 | 0.44 ± 0.04 |
| 1170 ATU | 117 days | 2.2 ± 0.13 | 1.6 ± 0.15 | 0.64 ± 0.02 |
Spectral Absorbance of Visual Pigments in Juvenile Halibut
Table 2: Visual pigment diversity in post-metamorphic Atlantic halibut as determined by MSP. Data adapted from [2].
| Photoreceptor Type | Visual Pigment Class | Mean Wavelength of Maximum Absorbance (λmax, nm) |
|---|---|---|
| Rod | RH1 | 491 |
| Single Cone | SWS (Short-Wavelength Sensitive) | 431, 457 |
| Single/Double Cone | RH2 (Middle-Wavelength Sensitive) | 500, 514, 527 |
| Double Cone | LWS (Long-Wavelength Sensitive) | 550 |
Experimental Protocols
Protocol: Microspectrophotometry (MSP) of Retinal Photoreceptors
Principle: MSP measures the absorbance spectrum of visual pigments within individual photoreceptor cells by passing a monochromatic beam of light through the outer segment and comparing it to a reference beam [2] [12].
Materials:
- Fresh or dark-adapted frozen retina from juvenile Atlantic halibut
- Microspectrophotometer
- Microscope with infrared-sensitive camera for cell manipulation
- Physiological saline solution
- Glass slides and cover slips
- Tools for fine dissection
Procedure:
- Tissue Preparation: Under dim red light, enucleate the eye and hemisect it. Remove the retina and place it in a drop of physiological saline on a glass slide. Gently tease the retinal tissue apart using fine needles to liberate photoreceptor cells. Apply a cover slip [2] [12].
- Instrument Calibration: Calibrate the MSP with a neutral density filter and ensure the baseline spectrum is stable.
- Cell Selection & Alignment: View the sample using infrared illumination. Identify and align a single photoreceptor outer segment in the measuring beam.
- Spectral Scanning: Scan the monochromatic light across the relevant wavelength range (e.g., 350-650 nm). Record the transmitted light intensity through the cell at each wavelength.
- Data Processing: For each cell, calculate absorbance as a function of wavelength. Fit the resulting absorbance spectrum with a standard visual pigment template to determine the wavelength of maximum absorbance (λmax) [2].
- Chromophore Identification: To confirm the type of chromophore (A1 or A2), expose the pigment to deep red light, which will bleach pigments based on their chromophore type, and note the spectral changes in the difference spectrum [12].
Protocol: Retinal Histology and Photoreceptor Mosaic Analysis
Principle: This protocol details the preparation of retinal cross-sections to visualize and characterize the spatial organization of photoreceptor mosaics at different developmental stages.
Materials:
- Fixative (e.g., 4% Paraformaldehyde in 0.1M Phosphate Buffer)
- Cryostat or microtome
- SuperFrost Plus glass slides
- Stains (e.g., Toluidine blue) or dyes for differential interference contrast (DIC) microscopy
- Fluorescence microscope
Procedure:
- Fixation: Immerse dissected eye cups immediately in fixative for 24 hours.
- Tissue Processing: Dehydrate the fixed tissue through a graded ethanol series, clear it, and embed it in paraffin or a resin for high-resolution sectioning. Alternatively, for cryosectioning, immerse the tissue in a sucrose solution before embedding in OCT compound [2] [27].
- Sectioning: Cut thin sections (1-15 μm thickness) using a microtome or cryostat and mount them on glass slides.
- Staining & Imaging: Stain sections with a histological stain like Toluidine blue to enhance contrast or use DIC microscopy to visualize unstained photoreceptor inner and outer segments [2].
- Mosaic Analysis: Image the photoreceptor layer at high magnification across different retinal regions (dorsal, ventral, nasal, temporal). Identify and classify photoreceptors as single, double, or triple cones. Map their spatial arrangement to classify the mosaic type (e.g., honeycomb vs. square) [2].
Visualizing the Experimental Workflow
The following diagram illustrates the integrated methodology from tissue preparation to data analysis, as applied in the halibut case study.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential materials and reagents for photoreceptor and visual pigment research.
| Reagent / Material | Function / Application |
|---|---|
| 11-cis Retinal Chromophore | The light-sensitive component of visual pigments; used to regenerate pigments in vitro [12]. |
| Paraformaldehyde (4% in Buffer) | Fixative for preserving retinal tissue morphology for histological analysis [2] [27]. |
| Physiological Saline Solution | Isotonic solution for maintaining photoreceptor viability during dissection and MSP preparation [2]. |
| OCT Compound | Embedding medium for freezing and stabilizing retinal tissue for cryosectioning [27]. |
| Digoxigenin (DIG)-labeled cRNA Probes | For in situ hybridization to localize specific opsin mRNA expression within retinal sections [27]. |
| Anti-Rhodopsin / Anti-Opsin Antibodies | For immunohistochemical labeling of specific photoreceptor types (rods/cones) in retinal sections [27]. |
| RNA Preservation Reagent (e.g., RNAlater) | Stabilizes RNA for subsequent molecular analysis of opsin gene expression via qPCR or RNA-Seq [12]. |
This application note provides a detailed protocol for the microspectrophotometric (MSP) analysis of visual pigments and oil droplets in the avian retina, using the wedge-tailed shearwater (Puffinus pacificus) as a marine species case study [18]. The methodology outlined enables the quantitative characterization of photoreceptor spectral sensitivities and the investigation of topographic variations across the retina. The data and procedures are instrumental for researchers studying visual ecology, sensory adaptation, and the physiological impacts of light pollution on wildlife [28].
The following tables consolidate the key spectral absorbance data obtained from the wedge-tailed shearwater retina via MSP.
Table 1: Visual Pigment and Oil Droplet Characteristics in Peripheral Retina
| Photoreceptor Type | Visual Pigment λmax (nm) | Oil Droplet Type | Oil Droplet λcut (nm) |
|---|---|---|---|
| Rod | 502 | Not Applicable | Not Applicable |
| VS Single Cone | 406 | T-type (Transparent) | >370 [18] |
| SWS Single Cone | 450 | C-type | 445 [18] |
| MWS Single Cone | 503 | Y-type | 506 [18] |
| LWS Single Cone | 566 | R-type | 562 [18] |
| Double Cone (Principal) | 566 | P-type | 413 [18] |
| Double Cone (Accessory) | 566 | None | Not Applicable [18] |
Table 2: Photoreceptor Density and Acuity Adaptations
| Retinal Region | Photoreceptor Characteristics | Adaptive Significance |
|---|---|---|
| Peripheral Retina | Heavily pigmented oil droplets [18] | Enhanced colour discrimination and glare reduction [29] |
| Central Horizontal Streak | Colourless/less pigmented oil droplets; narrower photoreceptors [18] | Increased photon capture for high visual acuity [18] |
Detailed Experimental Protocol: Microspectrophotometry
1.0 Retinal Tissue Preparation
- 1.1 Dissection and Isolation: Under dim red light, suitable for dark-adapting the retina, enucleate the eye and hemisect it. Remove the retina and immerse it in a physiological saline solution (e.g., phosphate-buffered saline). For MSP, small pieces of the retina may be gently mashed between a microscope slide and coverslip to separate the photoreceptor cells [30].
- 1.2 Tissue Fixation (Alternative for Histology): For morphological studies, fix retinal sections in a solution of 1% paraformaldehyde and 1.6% glutaraldehyde in a 0.1M phosphate buffer (pH 7.4) for 2 hours, followed by post-fixation in 2% osmium tetroxide [31]. Dehydrate through a graded ethanol series and embed in epoxy resin for sectioning.
2.0 Microspectrophotometric Analysis
- 2.1 Instrument Calibration: Calibrate the microspectrophotometer using a neutral density filter and a known standard for wavelength accuracy before measurement sessions.
- 2.2 Absorbance Measurements:
- 2.2.1 Visual Pigments: Position a monochromatic beam of light (e.g., 1 x 2 µm) onto the outer segment of a photoreceptor. Record the transmitted light across the spectrum (e.g., 350-750 nm) to generate an absorbance spectrum. The wavelength of maximum absorbance (λmax) is determined by fitting a visual pigment template curve to the raw data [18] [30].
- 2.2.2 Oil Droplets: Similarly, position the beam on an individual oil droplet and measure its spectral transmittance. The cut-off wavelength (λcut) is defined as the wavelength at which transmittance reaches 50% [18].
3.0 Topographic Mapping
- 3.1 Retinal Mapping: Systematically analyze the retina from central to peripheral regions (e.g., temporal, nasal, dorsal, ventral) [31]. For each zone, identify and record the types and relative densities of photoreceptors and their associated oil droplets.
- 3.2 Data Correlation: Correlate the MSP data with retinal topography to identify regional specializations, such as the horizontal streak of high photoreceptor density observed in the shearwater [18].
Visual System Workflow and Structure
The following diagram illustrates the experimental workflow for MSP analysis and the resulting organization of photoreceptors in the shearwater retina.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Retinal MSP Analysis
| Item | Function/Description | Application Note |
|---|---|---|
| Microspectrophotometer | Instrument for measuring absorbance spectra of microscopic structures [18]. | Requires a UV-vis light source and a sensitive detector for measuring single cells. |
| Physiological Saline | Buffer solution to maintain tissue viability during dissection. | Phosphate-buffered saline (PBS) at pH 7.4 is commonly used. |
| Fixative Solution | Chemical mixture to preserve retinal tissue for histology. | A common formulation is 1% paraformaldehyde, 1.6% glutaraldehyde in 0.1M phosphate buffer [31]. |
| Embedding Resin | Medium for tissue support for thin-sectioning. | Epoxy resins (e.g., Epon-812) are standard for electron microscopy studies [31]. |
| Visual Pigment Template | Mathematical model for determining λmax from raw data. | Template curves derived from known visual pigments are fitted to MSP absorbance data [30]. |
This application note provides a detailed protocol for the investigation of visual pigments in marine species using microspectrophotometry (MSP). Focusing specifically on the calculation of maximum absorbance wavelength (λmax) and the analysis of absorbance spectra, we outline standardized methodologies for obtaining reliable spectral data from delicate marine organisms. The techniques described herein enable researchers to correlate visual pigment characteristics with ecological adaptations in varied aquatic light environments. Our comprehensive framework covers experimental design, data acquisition, and interpretation protocols tailored for marine research applications where sample integrity is paramount and ship-based analyses are often necessary.
Visual pigments, composed of an opsin protein bound to a chromophore, are fundamental to phototransduction and ecological adaptation in marine species [32]. Microspectrophotometry provides the unique capability to measure absorbance spectra from individual photoreceptor cells, offering critical insights into visual function without requiring extensive tissue samples [5] [4]. For marine researchers, this technique is particularly valuable when studying fragile organisms that cannot survive transport to shore-based laboratories, enabling direct spectral measurements aboard research vessels [4].
The spectral sensitivity of visual pigments is characterized by their wavelength of maximum absorbance (λmax), a key parameter reflecting molecular structure and environmental adaptation [33] [32]. Accurate determination of λmax and proper analysis of spectral band shapes enables researchers to understand visual adaptation mechanisms in marine environments, where light transmission properties vary significantly with depth, water clarity, and geographic location [2] [32]. This protocol establishes standardized methodologies for spectral data interpretation specifically within the context of marine visual ecology.
Theoretical Foundations of Spectral Analysis
Visual Pigment Spectral Characteristics
Visual pigment absorbance spectra consist of multiple bands, with the principal α-band determining spectral sensitivity and secondary β-bands contributing to ultraviolet wavelength sensitivity [33]. The α-band is optimally described by mathematical templates that calculate normalized absorbance across the spectrum using λmax as the primary parameter [33]. These templates account for the invariant shape of visual pigment spectra when plotted on an inverse wavelength scale, allowing accurate prediction of spectral properties across diverse marine taxa.
For marine species, spectral tuning occurs primarily through amino acid replacements at critical positions in the opsin protein that alter the interaction with the chromophore within the retinal-binding pocket [32]. Marine environments exert distinct selective pressures on visual systems, with deep-water habitats dominated by narrow-band blue light around 475 nm, while shallow aquatic environments contain a broader spectrum similar to terrestrial light conditions [32]. These environmental differences drive evolutionary adaptations reflected in λmax values of marine visual pigments.
Mathematical Templates for λmax Determination
Two primary mathematical frameworks exist for describing visual pigment absorbance spectra, both utilizing λmax as the single essential parameter:
Stavenga-Smolka-Pirih (SSH) Template [33] [34]: This model employs a simple exponential function of the form:
where x = log10(λ/λmax), with parameters a = 380 and b = 6.09 for vitamin A1-based pigments typically found in marine crustaceans and fishes.
Govardovskii-Towner-Rohlich (Gov) Template [33]: This more recent template utilizes a seven-parameter equation:
where x = (λ/λmax) - 1 with predefined coefficients A, B, C, D and additional width parameters.
Both templates accurately describe the α-band of rhodopsins with λmax > 400 nm across approximately three log units of absorbance, with the Govardovskii template providing superior performance at very low absorbances (< 10⁻³) and in the ultraviolet wavelength range [33].
Experimental Protocols
Microspectrophotometry Setup for Marine Samples
Equipment Configuration: The fundamental MSP system for marine research replaces conventional single-channel photomultipliers with multichannel detectors, eliminating scanning-related artifacts caused by mechanical vibrations—particularly problematic in shipboard environments [5] [4]. This configuration enables simultaneous measurement across multiple wavelengths, crucial for analyzing photolabile visual pigments from marine organisms. Essential components include: a stable broadband light source, monochromator or interference filters, microscope with UV-transparent optics, sample chamber with temperature control, multichannel array detector (CCD or photodiode array), and vibration-damping platform for ship-based operations.
Marine Sample Preparation:
- Tissue Isolation: Dissect eyes under dim red light (λ > 650 nm) to prevent pigment bleaching. For ship-based work, perform dissections in chilled, oxygenated artificial seawater matching the species' natural habitat.
- Photoreceptor Isolation: Gently tease retinal tissue apart using fine needles to isolate individual photoreceptors or intact rod outer segments. For cone-dominant species like fully aquatic sea snakes, isolate entire cone cells [32].
- Sample Stabilization: Suspend photoreceptors in appropriate marine-balanced saline containing 5-10% sucrose or Ficoll for osmotic balance. Transfer 5-10 μL aliquots to quartz slide for measurement.
- Photopigment Regeneration: For pigment characterization, pre-incubate samples with 11-cis retinal (25-50 μM) for 4-12 hours at 4°C to ensure complete chromophore binding.
Absorbance Spectrum Measurement Protocol
System Calibration:
- Record baseline spectrum with clear reference area adjacent to sample
- Wavelength calibration using holmium oxide or didymium glass standards
- Intensity normalization across spectral range using neutral density filter
Spectral Acquisition:
- Position isolated photoreceptor in measurement beam (typically 1-2 μm spot size)
- Record initial sample spectrum (I) and reference spectrum (I₀)
- Calculate absorbance as A = -log(I/I₀)
- Repeat measurements at multiple orientations for dichroic samples
Difference Spectroscopy:
- Record pre-bleach absorbance spectrum
- Expose sample to intense white light (30-60 seconds) to photobleach visual pigment
- Record post-bleach absorbance spectrum
- Calculate difference spectrum (pre-bleach minus post-bleach)
- For marine invertebrate pigments, establish photoequilibrium using monochromatic illumination to assess metarhodopsin spectra [33]
Data Quality Validation:
- Accept spectra with minimum peak absorbance > 0.02 and maximum < 1.2
- Discard spectra showing obvious photobleaching during measurement
- Require consistent baseline in long-wavelength region (> 650 nm)
- Verify symmetrical α-band shape without secondary contamination
λmax Calculation Methodology
Raw Data Preprocessing:
- Apply digital smoothing algorithms (e.g., Savitzky-Golay filter) to reduce noise without distorting spectral shape [5]
- Subtract long-wavelength baseline (620-650 nm) to correct for light scattering
- Normalize spectrum to maximum absorbance = 1.0
Template Fitting Procedure:
- Select appropriate template based on chromophore type (A1 or A2)
- Implement iterative least-squares fitting algorithm to determine λmax
- For marine vertebrate pigments, use Govardovskii template with A1 coefficients for most accurate λmax determination [33]
- Assess fit quality using residual sum of squares and visual inspection
Validation and Reporting:
- Calculate standard error of λmax estimate from fitting procedure
- Report mean λmax ± SEM from multiple cells (typically n ≥ 5-10)
- Include goodness-of-fit metrics (R²) for individual spectra
- Document any spectral contamination or unusual band shapes
Data Presentation
Spectral Characteristics of Marine Visual Pigments
Table 1: Experimentally Determined λmax Values from Marine Species
| Species | Common Name | Photoreceptor Type | Visual Pigment | λmax (nm) | Method | Citation |
|---|---|---|---|---|---|---|
| Euphausia pacifica | North Pacific krill | Rhabdomeric | Rhodopsin | 483 | MSP | [4] |
| Euphausia pacifica | North Pacific krill | Rhabdomeric | Metarhodopsin | 489 | MSP | [4] |
| Hippoglossus hippoglossus | Atlantic halibut | Rod | Rhodopsin | 491 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Single cone | S(431) | 431 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Single cone | S(457) | 457 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Double cone | M(500) | 500 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Double cone | M(514) | 514 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Double cone | M(527) | 527 | MSP | [2] |
| Hippoglossus hippoglossus | Atlantic halibut | Double cone | L(550) | 550 | MSP | [2] |
| Callinectes sapidus | Blue crab | Rhabdom | Rhodopsin | ~500 (in vivo variation) | MSP | [5] |
Table 2: Template Function Parameters for Visual Pigment Spectra
| Template | Chromophore Type | α-band Parameters | β-band Parameters | Applicability |
|---|---|---|---|---|
| Stavenga et al. (1993) | A1 (marine typical) | a = 380, b = 6.09 | Aβ = 0.29, λβ = 350 nm | λmax > 400 nm, 3 log units |
| Govardovskii et al. (2000) | A1 (marine typical) | A=69.7, B=28, C=-14.9, D=0.674 | Aβ=0.26, λβ=189+0.315λmax | Full range, especially <10⁻³ absorbance |
| Mansfield & MacNichol | A1 & A2 | Wavelength-scale invariant | Not specified | Historical context |
Spectral Tuning in Marine Species
Table 3: Spectral Tuning Mechanisms in Marine Vertebrates
| Species Group | Opsin Type | Spectral Tuning Sites | Spectral Effect | Environmental Adaptation |
|---|---|---|---|---|
| Sea Snakes (Hydrophiini) | LWS | 164, 181, 269, 292 | Stepwise blue shift | Deep/open water (blue light) |
| Sea Snakes (Hydrophiini) | RH1 | 292 | Blue shift | Deep water environment |
| Sea Snakes (Hydrophiini) | SWS1 | 82 (Phe/Tyr polymorphism) | UV/blue sensitivity | Maintained polymorphism |
| Cetaceans (deep-diving) | RH1 | 83N, 292S, 299A | ~479 nm (blue shift) | Deep water foraging |
| Cetaceans (shallow) | RH1 | 83D, 292A, 299S | ~500 nm | Shallow water vision |
| Atlantic Halibut | Multiple cone opsins | Not specified | 431, 457, 500, 514, 527, 550 nm | Benthic lifestyle multi-mosaic |
Visualization of Methodologies
Microspectrophotometry Workflow
Spectral Data Analysis Procedure
The Scientist's Toolkit
Table 4: Essential Research Reagents and Materials for Marine Visual Pigment Analysis
| Item | Specification | Application | Notes |
|---|---|---|---|
| Artificial Seawater | Marine species-specific formulation | Sample dissection and maintenance | Match natural osmolarity and ion composition |
| 11-cis Retinal | 25-50 μM in ethanol | Visual pigment regeneration | Essential for complete chromophore binding |
| Sucrose/Ficoll | 5-10% in balanced saline | Osmotic stabilization during measurement | Prevents photoreceptor damage |
| Quartz Microscope Slides | UV-transparent | Sample mounting for MSP | Essential for UV spectrum measurements |
| Vibration-damping Platform | Ship-compatible | At-sea measurements | Critical for ship-based MSP [4] |
| Multichannel Detector | CCD or photodiode array | Simultaneous multi-wavelength detection | Eliminates scanning artifacts [5] |
| Interference Filters | Monochromatic illumination | Difference spectroscopy | Establish photoequilibrium states |
| Holmium Oxide Standard | NIST-traceable | Wavelength calibration | Essential for accurate λmax determination |
The methodologies outlined in this application note provide a comprehensive framework for accurate determination of λmax and analysis of visual pigment spectra in marine species. The implementation of multichannel microspectrophotometry has revolutionized ship-based analyses of fragile marine organisms, enabling direct investigation of visual adaptations to aquatic environments without sample degradation during transport [4]. Through standardized protocols for spectral measurement, template-based λmax calculation, and data validation, researchers can obtain reliable, reproducible results that illuminate the relationship between visual function and ecology in marine systems.
The mathematical templates developed by Stavenga et al. [34] and Govardovskii et al. [33] provide robust tools for spectral analysis, with each offering particular advantages for different applications in marine research. When combined with contemporary molecular techniques including opsin sequencing and phylogenetic analysis, microspectrophotometry forms an essential component of integrative approaches to understanding visual adaptation in marine environments [2] [32]. The continued refinement of these methodologies will further enhance our ability to decipher the remarkable visual adaptations that enable marine species to thrive in diverse light environments.
Navigating Technical Challenges in Marine Microspectrophotometry
Mitigating Signal-to-Noise Issues in Low-Pigment Concentrations
Microspectrophotometry (MSP) serves as an indispensable technique in visual ecology, enabling researchers to measure the absorption properties of visual pigments directly from individual photoreceptor cells of marine species [35]. This application is particularly valuable for studying the spectral adaptations of fish and other marine organisms to their specific photic environments. However, the successful application of MSP to marine research faces a significant challenge: mitigating signal-to-noise ratio (SNR) issues that arise when analyzing microscopic samples with low pigment concentrations. These SNR constraints can fundamentally limit the reliability of spectral data and subsequent ecological interpretations. The technical foundation of MSP involves passing a narrow beam of light (approximately 2×2 μm in cross-section) through a single cell and measuring the amount of light transmitted at each wavelength [35]. When working with marine species that may have thin photoreceptors or sparse pigment packing, the resulting weak signals require sophisticated optimization strategies to extract meaningful data. This application note provides a structured framework for addressing these SNR challenges through optimized methodologies and specialized reagent solutions.
Technical Principles and SNR Challenges in MSP
Fundamental MSP Configuration for Visual-Pigment Analysis
In marine visual ecology, MSP is uniquely capable of measuring spectral absorption properties of visual pigments in situ from individual retinal photoreceptors [35]. The instrument configuration typically integrates microscope optics with a single-beam-configuration spectrophotometer, linked to computer systems for operation and spectral comparison [35]. This setup allows researchers to work with the microscopic photoreceptors common in marine species. The measurement principle relies on the fact that visual pigment color is determined by which wavelengths of the visible electromagnetic spectrum are absorbed by the chromophore-retinal complex [35]. For a marine fish adapted to blue-shifted oceanic environments, the absorption spectrum would peak at shorter wavelengths compared to a species inhabiting turbid coastal waters.
The analysis of marine visual pigments extends beyond simple absorption peaks to encompass the photochemical stability and thermal noise properties of the pigment molecules. Visual pigments can undergo spontaneous activation driven by thermal energy, generating noise that interferes with accurate detection of real-light signals [36]. This thermal noise manifests as electrical events indistinguishable from those triggered by absorbed photons, creating a "dark light" that sets the fundamental detection threshold for vision [36]. In MSP measurements of low-concentration pigments, this molecular-level noise combines with optical and electronic noise sources to degrade SNR.
Critical SNR Challenges in Marine Pigment Research
Table 1: Primary Signal-to-Noise Challenges in Marine Visual-Pigment MSP
| Challenge Category | Specific Issue | Impact on SNR |
|---|---|---|
| Sample Characteristics | Low pigment density in slender photoreceptors | Reduced signal amplitude, increased relative noise |
| Light scattering in ocular media | Decreased transmission signal, elevated background | |
| Pigment bleaching during measurement | Signal degradation over time | |
| Instrument Limitations | Stray light interference | Increased background noise, reduced contrast |
| Detector electronic noise | Obscures weak absorption signals | |
| Limited photon flux through micro-apertures | Poor counting statistics | |
| Environmental Factors | Thermal activation of pigments [36] | Increased molecular noise floor |
| Chromophore-exchange in cone pigments [36] | Signal instability during measurement |
Marine researchers face particular difficulties because many fish species possess multiple visual pigments adapted to specific depth ranges and water compositions, often at low concentrations that challenge detection limits [37]. The spectral properties of visual pigments in Baltic Sea fishes, for example, show adaptations to the brackish light environment with rod absorbance spectra exhibiting wavelength-shifting of several nanometers compared to their marine counterparts [37]. Detecting these subtle but ecologically significant spectral differences demands optimized SNR conditions. Furthermore, the spontaneous thermal activation of pigments varies with the openness of the chromophore-binding pocket, with cone pigments typically being approximately 25-fold noisier than rod pigments of the same λmax [36]. This molecular property directly impacts the achievable SNR in MSP measurements of different photoreceptor types.
Optimized Experimental Protocols for Enhanced SNR
Sample Preparation and Stabilization Methods
Protocol 3.1: Retinal Tissue Preparation for Marine Species
- Dark Adaptation: Maintain experimental marine specimens in complete darkness for 2-4 hours prior to retinal extraction to ensure visual pigments are fully regenerated.
- Retinal Dissection: Perform dissection under infrared illumination using image converters to prevent pigment bleaching. Use chilled, oxygenated physiological saline appropriate for the species being studied.
- Photoreceptor Isolation: Gently tease apart retinal tissue using vibratome sectioning or enzymatic treatment (e.g., mild papain digestion) to isolate individual photoreceptors while maintaining structural integrity.
- Sample Mounting: Transfer isolated photoreceptors to quartz microscope slides using glycerin as a mountant [35]. For UV measurements, quartz coverslips are essential as glass absorbs ultraviolet radiation.
- Orientation Control: Carefully orient reference and suspect fibers in the same way for analysis to avoid pleochroic effects that can introduce spectral artifacts [35].
Protocol 3.2: Pigment Stability Enhancement
- Chromophore Stabilization: For cone pigments prone to spontaneous dissociation, consider adding chromophore-binding proteins (e.g., CRALBP) to the mounting medium to reduce dark exchange [38].
- Temperature Control: Maintain samples at consistent temperatures (typically 15-20°C for cold-water species) throughout measurement to minimize thermal activation noise [36].
- Antioxidant Supplementation: Add antioxidant compounds (e.g., ascorbate) to mounting media to reduce oxidative damage to unsaturated retinal chromophores during measurement.
Instrument Configuration and Measurement Optimization
Protocol 3.3: MSP Instrument Calibration for Low-Pigment Work
- Aperture Optimization: Precisely control analysis area using instrument apertures to match photoreceptor dimensions while excluding background [39]. Typical apertures of 2×2 μm provide optimal balance between signal collection and spatial resolution.
- Spectral Range Configuration: Utilize quartz optics to extend measurements down to ~200 nm for detecting UV-absorbing compounds [39]. Combine tungsten halogen, xenon, and deuterium illuminators to cover UV through near-infrared spectral regions as needed.
- Reference Calibration: Collect reference spectra from clean, pigment-free areas of the sample slide immediately before and after sample measurements to account for instrumental drift.
- Background Subtraction: Implement synchronized background subtraction protocols using adjacent blank areas to minimize scattering and mounting medium artifacts.
Protocol 3.4: Enhanced SNR Measurement Protocol
- Signal Averaging: Accumulate multiple scans (typically 8-16) for each measurement location to improve counting statistics while monitoring for bleaching effects.
- Spectral Binning: Apply moderate spectral binning (2-4 nm) when high spectral resolution is not critical to increase signal at each data point.
- Dynamic Range Optimization: Adjust integration times and amplifier gains to utilize the full dynamic range of the detection system without saturation.
- Polarization Control: Use plain polarized light to study pleochroism when relevant, but employ unpolarized light for standard comparisons to avoid orientation-dependent artifacts [39].
Reagent Solutions and Materials for Marine Visual Pigment MSP
Table 2: Essential Research Reagent Solutions for Marine Visual Pigment Analysis
| Reagent/Material | Specification | Functional Application |
|---|---|---|
| Mounting Medium | Glycerin, spectrophotometric grade | Maintains hydration and optical clarity for transmission measurements [35] |
| Physiological Saline | Species-specific formulation with balanced ions | Maintains photoreceptor viability during dissection and preparation |
| Chromophore Analogs | 9-demethyl retinal, 11-cis retinal analogs [38] | Investigate opsin-chromophore interactions and pigment regeneration kinetics |
| Binding Proteins | CRALBP, arrestin variants [38] | Stabilize chromophore binding, reduce spontaneous exchange in cone pigments |
| Antioxidants | Ascorbate, α-tocopherol | Protect unsaturated chromophores from oxidative damage during measurement |
| Fixatives | Low-concentration glutaraldehyde (0.1-0.5%) | Optional stabilization for difficult samples; may alter spectral properties |
Data Analysis and Interpretation Framework
Spectral Processing and Noise Reduction Techniques
Following optimized data collection, appropriate spectral processing is essential for extracting meaningful information from low-signal MSP measurements. The first derivative transformation provides a valuable mathematical approach for exacerbating subtle spectral differences [35]. This function, calculated as ∂γ/∂λ = (γi+1 - γi)/(λi+1 - λi), where γ represents absorption values at wavelength intervals λ, enables visualization of the rate of change in spectral gradients [35]. However, this technique must be used cautiously as it can potentially lead to false exclusions in cases with high intra-sample variation [35]. For marine species with potentially high individual variation, such as populations adapting to different photic environments, validation against raw absorption spectra is essential.
Complementary to derivative analysis, digital filtering approaches can enhance SNR during post-processing. Savitzky-Golay smoothing preserves spectral features while reducing high-frequency noise, particularly valuable for the typically noisy signals from low-concentration pigments. Additionally, multivariate analysis techniques such as principal component analysis can help distinguish true spectral features from noise in datasets from multiple photoreceptors. When comparing spectra from marine populations, researchers should consider both the wavelength positions and intensities of absorption maxima and minima, along with the appearance and positions of individual spectral features such as peak shoulders or points of inflection [35].
Interpretation Guidelines for Marine Visual Ecology
In the context of marine visual ecology, MSP data interpretation must extend beyond technical spectral matching to consider ecological relevance. The study of Baltic Sea fishes provides an excellent example, where spectral differences in rod visual pigments between marine and brackish-water populations were interpreted as adaptations for improved quantum catch and enhanced signal-to-noise ratio in the specific Baltic light environment [37]. When λmax values fall outside theoretically optimal ranges for a water type, as observed for sandeels in the Baltic Sea, researchers should consider behavioral explanations such as activity patterns restricted to bright light conditions [37].
For rigorous comparison, establish quantitative matching criteria that account for both spectral shape and amplitude variations. The criterion should consider the natural variation within a population, as marine individuals from different habitats may exhibit significant spectral differences reflecting local adaptations. Particularly for marine species with high intra-retinal variation, ensure that reference samples adequately represent the natural diversity present in the population to avoid false exclusions. Document both the mean spectral properties and the range of variation observed within samples, as this information is crucial for interpreting ecological adaptations and phylogenetic relationships.
Workflow and Phototransduction Pathway Visualization
Experimental Workflow for Marine Visual Pigment MSP
Diagram Title: Marine Visual Pigment MSP Workflow
Diagram Title: Phototransduction Pathway and Noise
Mitigating signal-to-noise issues in microspectrophotometry of low-concentration visual pigments requires an integrated approach spanning sample preparation, instrument optimization, and data analysis. The protocols and methodologies presented here provide a systematic framework for enhancing SNR in marine visual pigment research, enabling more reliable characterization of spectral adaptations in aquatic environments. By addressing both technical and biological sources of noise, researchers can extract meaningful data even from challenging samples with low pigment densities. The specialized reagent solutions and optimized workflows specifically support investigations into marine visual ecology, where subtle spectral differences often reflect significant adaptations to photic environment. As research in this field advances, these methodologies will facilitate deeper understanding of the relationship between visual pigment properties and ecological niche specialization in marine species.
This application note establishes a comprehensive framework for validating functional vision in marine species, bridging the gap between laboratory-based visual pigment characterization and ecologically relevant visual performance. We provide detailed protocols for integrating microspectrophotometry (MSP) with behavioral and environmental assessments to determine how visual capabilities contribute to survival, feeding, and reproductive success in marine environments. This approach moves beyond merely confirming the presence of visual pigments to understanding their functional significance within complex ecological contexts.
Traditional visual pigment analysis via microspectrophotometry has primarily focused on characterizing absorption spectra and identifying photopigment types in marine organisms [40] [41]. While these data provide essential foundation information about visual potential, they reveal limited information about how these capabilities are actually deployed in ecological contexts. The distinction between visual function (performance of visual system components) and functional vision (visual task-related ability in real-world scenarios) is crucial for ecological validation [42].
Marine organisms inhabit complex visual environments where factors such as light attenuation, spectral shifting, turbidity, and motion detection create evolutionary pressures that shape visual system functionality. For example, studies of crustacean visual systems have revealed multiple visual pigments potentially supporting chromatic discrimination and adapted to specific photic environments [40] [41]. This protocol series provides methodologies to quantitatively connect laboratory measurements with ecological performance through standardized tests, environmental simulations, and integrated data interpretation frameworks.
Theoretical Foundation: Visual Assessment Hierarchy
Conceptual Framework
The validation of functional vision requires a hierarchical approach that connects molecular, physiological, behavioral, and ecological levels of analysis. This framework ensures that measurements at each level inform our understanding of functionality at subsequent levels, creating a comprehensive profile of visual ecology.
Key Definitions and Terminology
Table 1: Standardized Terminology for Functional Vision Research
| Term | Definition | Application in Marine Context |
|---|---|---|
| Visual Function | Performance of components of the visual system under controlled conditions [42] | Spectral sensitivity measurements, visual acuity thresholds, contrast sensitivity |
| Functional Vision | Visual task-related ability under real-world scenarios and ecological conditions [42] | Prey detection in turbid water, predator avoidance, mate selection under ambient light |
| Visual Pigment | Light-sensitive molecules in photoreceptors that initiate visual transduction | Rhodopsin, metarhodopsin characterized by MSP [40] [41] |
| Ecological Contribution | The role of visual capabilities in survival, reproduction, and fitness | Determining how spectral tuning matches habitat photic characteristics |
Integrated Methodological Approaches
Protocol 1: Ship-Based Microspectrophotometry for Marine Species
Purpose and Applications
This protocol enables direct characterization of visual pigments from fragile marine organisms immediately after collection, eliminating artifacts associated with transport to shore-based laboratories and preserving physiological integrity of visual tissues [40] [4].
Experimental Workflow
Technical Specifications
- System Configuration: Multichannel detector replacing conventional photomultiplier tubes
- Spectral Range: 350-700 nm (adaptable for specific marine environments)
- Spectral Resolution: 2-5 nm depending on grating and slit configuration
- Light Source: Stable quartz halogen with monochromator for test beams
- Reference Channel: Continuous monitoring of source stability
- Vibration Mitigation: Ship-optimized mounting system to eliminate mechanical artifacts
Data Processing and Analysis
- Raw Absorbance Calculation:
- A = -log₁₀(Sample transmission / Reference transmission)
- Baseline Correction:
- Subtract absorbance in non-absorbing spectral regions
- Difference Spectra Generation:
- Compute light-adapted minus dark-adapted spectra
- Peak Sensitivity Determination (λmax):
- Fit difference spectra with visual pigment template functions
- Photoproduct Identification:
- Characterize metarhodopsin and other stable photoproducts
Protocol 2: Ecological Contribution Assay Development
Behavioral Integration Framework
This protocol links MSP-derived visual capabilities with ecologically relevant behaviors through controlled laboratory assays that simulate key environmental challenges.
Table 2: Ecological Contribution Assessment Matrix
| Ecological Challenge | Laboratory Assay | Performance Metrics | MSP Correlation |
|---|---|---|---|
| Prey Detection | Moving target discrimination in varying turbidity | Detection distance, success rate, pursuit initiation | Spectral sensitivity match to prey contrast |
| Predator Avoidance | Looming stimulus response | Reaction distance, escape trajectory, response latency | Motion sensitivity and temporal resolution |
| Mate Selection | Conspecific cue discrimination | Choice preference, display response, courtship intensity | Color vision capabilities and UV sensitivity |
| Habitat Navigation | Spatial maze with light cues | Navigation efficiency, learning rate, cue utilization | Scotopic vs. photopic sensitivity thresholds |
Environmental Simulation Parameters
- Light Field Control: Spectral composition, intensity, and angular distribution matching depth-specific conditions
- Water Quality Parameters: Turbidity, chlorophyll content, CDOM concentration
- Background Visual Noise: Natural substrate patterns, plankton blooms, surface glitter patterns
- Stimulus Presentation: Computer-controlled displays with calibrated spectral output
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Functional Vision Research
| Item | Specification | Functional Application | Ecological Validation |
|---|---|---|---|
| Multichannel MSP System | Ship-deployable with vibration damping [40] [4] | Visual pigment characterization in fresh tissues | Direct correlation with collection environment conditions |
| Spectroradiometer | Underwater capable, 350-800 nm range | Habitat light field quantification | Determine spectral tuning to environmental cues |
| Environmental Simulation Chamber | Programmable LED arrays, turbidity control | Behavioral assay presentation | Ecologically relevant performance assessment |
| Visual Stimulation System | High refresh rate, calibrated spectral output | Visual capability testing | Link pigment properties to visual task performance |
| Tissue Preservation Media | Marine-specific osmolarity, antioxidant supplements | Maintain photoreceptor integrity during processing | Ensure physiological relevance of MSP measurements |
Data Presentation and Interpretation Framework
Quantitative Visualization Standards
All data presentation should follow established guidelines for scientific communication [43] [44]. Continuous data (e.g., spectral absorbance, contrast sensitivity) should be presented with distribution-preserving visualizations (scatterplots, box plots), while categorical data (e.g., behavioral choices) benefit from bar charts with appropriate error representation.
Color Contrast Requirements for Scientific Visualization
All experimental diagrams and data visualizations must meet minimum color contrast ratios of 4.5:1 for standard text and visual elements [45]. This ensures accessibility and accurate interpretation across varying display conditions and for researchers with color vision deficiencies. The specified color palette (#4285F4, #EA4335, #FBBC05, #34A853, #FFFFFF, #F1F3F4, #202124, #5F6368) provides sufficient luminance contrast when appropriately paired.
This integrated protocol series enables researchers to move beyond the mere presence of visual pigments to understanding their functional contribution to ecological success. By combining ship-based MSP with carefully designed ecological assays, researchers can establish causal links between visual system properties and fitness-relevant behaviors. The provided frameworks standardize terminology and methodology across the field of marine visual ecology, facilitating comparative studies and meta-analyses that advance our understanding of visual adaptation in aquatic environments.
Addressing Photopigment Bleaching and Sample Degradation
Photopigment bleaching and sample degradation represent significant challenges in microspectrophotometry (MSP) studies of marine species, potentially compromising data accuracy and reproducibility. These phenomena are particularly problematic when analyzing delicate visual pigments from marine organisms, which are often sensitive to light exposure and environmental fluctuations. This document provides detailed application notes and protocols to identify, mitigate, and account for these issues within the broader context of marine visual ecology research. The strategies outlined herein combine controlled experimental environments, advanced analytical techniques, and robust data normalization methods to enhance the reliability of MSP-derived data in studies of marine visual systems.
Background and Significance
Photopigment bleaching occurs when visual pigment molecules undergo molecular alterations upon light exposure, changing their spectral absorption properties. In marine research, this poses particular challenges as many species possess visual pigments adapted to low-light environments, making them exceptionally photosensitive. Sample degradation encompasses broader deterioration processes, including enzymatic activity, oxidative damage, and microbial contamination, all of which can alter pigment composition and spectral characteristics. These processes are especially pronounced in field-collected marine samples where immediate processing isn't always feasible. Understanding and controlling these artifacts is crucial for accurate characterization of visual pigments in marine species, which in turn informs studies of visual ecology, behavior, and sensory evolution in aquatic environments.
Quantitative Assessment of Photopigment Bleaching
Key Indicators and Metrics
The following table summarizes primary quantitative metrics for assessing photopigment bleaching and sample degradation in marine visual pigment studies:
Table 1: Quantitative Metrics for Assessing Photopigment Integrity
| Metric | Measurement Technique | Normal Range | Degradation Indicator | Marine Application Notes |
|---|---|---|---|---|
| Fv/Fm Ratio (Photosynthetic efficiency) | Pulse-amplitude modulation (PAM) fluorometry | 0.65-0.85 in healthy coral symbionts [46] | Values <0.45 indicate severe bleaching [46] | Critical for studies of coral visual systems; directly affects accessory pigment availability |
| Photopigment Content | Spectrophotometric analysis | Species-specific baselines | >40% reduction indicates significant bleaching [46] | Measure at λmax for marine visual pigments; compare to published standards |
| Spectral Shift Index | Microspectrophotometry | Stable λmax ± 2nm | Bathochromic shift >5nm suggests degradation | Particularly relevant for deep-sea species with UV-shifted visual pigments |
| DGCC Betaine Lipid Saturation | Liquid chromatography-mass spectrometry (LC-MS/MS) | Varies with thermal adaptation [47] | Reduced saturation correlates with bleaching susceptibility [47] | Biomarker for intergenerational bleaching resistance in marine organisms |
| Metabolome Diversity (Shannon Entropy) | Untargeted metabolomics | Stage-specific baselines [47] | Significant increase in bleached specimens [47] | Reflects broader biochemical disruptions in visual pigment pathways |
Advanced Monitoring Techniques
Modern approaches to monitoring pigment integrity extend beyond basic spectral measurements. Visible spectral imaging technology enables non-contact pigment analysis through wavelength-scan methods, spatial-scan methods, time-scan methods, and simultaneous acquisition methods [48]. These approaches are particularly valuable for rare marine specimens where minimal invasiveness is critical. Additionally, ATR-FTIR spectroscopy coupled with chemometric methods has demonstrated excellent capability in detecting molecular-level changes in biological samples, with high R² values (0.94-0.96) for degradation state prediction [49].
Experimental Protocols for Mitigating Bleaching and Degradation
Protocol 1: Probiotic Treatment for Coral Photopigment Preservation
This protocol adapts beneficial microorganism approaches for maintaining photopigment stability in coral holobionts, which can be extended to other marine symbiotic systems [46].
Materials:
- Native putative Beneficial Microorganisms for Corals (pBMCs) isolated from target species
- Marine Agar (MA) medium (e.g., Zobell 2216)
- Artificial seawater (Red Sea Salt recommended)
- Controlled temperature aquarium systems (±0.5°C precision)
Procedure:
- Isolate pBMCs from donor specimens using marine agar with 10⁻³ to 10⁻⁵ dilutions
- Culture pBMCs at 28°C for 16 hours; select colonies demonstrating catalase production
- Prepare pBMC consortium suspension in sterile saline solution (0.85%)
- For experimental specimens, administer pBMC suspension daily for 5-day acclimation
- Maintain control and treatment groups at stable temperature (26°C) and pH (8.1)
- Monitor photopigment content and Fv/Fm ratios daily for 7-14 days
- Process MSP samples under dim red light to prevent additional bleaching
Validation: Expect significantly higher photopigment content (p<0.05) and Fv/Fm ratios (≥0.65) in treated specimens compared to controls when exposed to sublethal thermal stress (30°C) [46].
Protocol 2: Metabolomic Stabilization for Marine Visual Tissue
This protocol utilizes metabolomic approaches to stabilize photopigments by maintaining biochemical integrity, based on intergenerational bleaching resistance studies [47].
Materials:
- Liquid nitrogen storage system
- LC-MS/MS system with C18 column
- Methanol, acetonitrile, and water (LC-MS grade)
- Cryogenic tissue pulverizer
Procedure:
- Rapidly excise ocular tissues under dim infrared illumination (>900nm)
- Immediately flash-freeze in liquid nitrogen (within 30 seconds of dissection)
- Store at -80°C until analysis (avoid repeated freeze-thaw cycles)
- For extraction, homogenize tissue in cold 80% methanol using cryogenic pulverization
- Centrifuge at 14,000g for 15 minutes at 4°C
- Collect supernatant and evaporate under nitrogen stream
- Reconstitute in 100μL methanol:water (1:1) for LC-MS/MS analysis
- Monitor DGCC betaine lipid saturation as key stability biomarker [47]
Validation: Successful stabilization should maintain DGCC lipid saturation states consistent with parent bleaching phenotype, with correlation coefficients ≥0.913 for melanin and ≥0.941 for hemoglobin compared to reference systems [47].
Protocol 3: Chemometric Assessment of Photopigment Degradation
This protocol adapts forensic bloodstain age determination methods for assessing photopigment degradation state in marine samples [49].
Materials:
- ATR-FTIR spectrometer with diamond crystal
- Unscrambler or similar chemometric software
- Partial Least Squares Regression (PLSR) algorithms
- Normal saline solution (0.9%)
Procedure:
- Prepare thin tissue sections (≤10μm) or pigment extracts on IR-transparent substrates
- Collect spectra in range 900-1800 cm⁻¹ at 4 cm⁻¹ resolution with 32 scans
- Apply preprocessing: baseline correction, vector normalization, multiplicative scatter correction
- Develop PLSR model using reference samples of known degradation states
- Validate model using 10-fold cross-validation with Venetian blinds procedure
- Apply model to unknown samples to quantify degradation index
- Establish acceptance criteria (e.g., RMSEP ≤5.83, R² ≥0.94, RPD ≥4.08) [49]
Validation: The model should successfully distinguish fresh (age ≤1 day) from degraded samples (age >1 day) with classification accuracy >95% based on PLS-DA modeling [49].
Research Reagent Solutions
Table 2: Essential Research Reagents for Addressing Photopigment Degradation
| Reagent/Category | Specific Examples | Function in Photopigment Research | Marine-Specific Considerations |
|---|---|---|---|
| Microbial Consortia | Pseudoalteromonas sp., Halomonas taeanensis, Cobetia marina-related species [46] | Reduce bleaching through antioxidant production and pathogen exclusion | Must be isolated from cognate marine host species or environment |
| Cryoprotectants | Dimethyl sulfoxide (DMSO), glycerol, trehalose | Stabilize pigment structures during frozen storage | Concentration must be optimized for marine tissue osmolarity |
| Antioxidants | Ascorbic acid, catalase, superoxide dismutase | Scramble reactive oxygen species that drive photobleaching | Catalase-producing isolates are particularly effective for coral systems [46] |
| Chelating Agents | EDTA, EGTA | Bind metal ions that catalyze oxidative degradation | Important for marine samples with high ambient ion concentrations |
| Metabolomic Standards | DGCC betaine lipids, deuterated amino acids, stable isotope labels | Quantitative references for pigment stability biomarkers | DGCC saturation state is key biomarker for thermal tolerance [47] |
| Spectroscopic Matrices | Potassium bromide, barium fluoride, diamond ATR crystals | Medium for IR and Raman analysis of pigment integrity | Diamond ATR crystals withstand marine salt residues [49] |
Workflow Visualization
Sample Integrity Workflow
Mitigation Strategy Map
Data Normalization and Reporting Standards
To ensure comparability across marine photopigment studies, implement rigorous normalization protocols that account for potential degradation artifacts. Report both raw and normalized spectra, explicitly documenting any correction factors applied. For MSP data, include reference measurements from standard pigments with known stability characteristics. When presenting results, clearly state the stabilization methods employed and provide degradation indices based on chemometric models (e.g., PLSR predictions with associated RMSEP values) [49]. For metabolomic data, report DGCC betaine lipid saturation states alongside traditional photopigment metrics, as these molecular features demonstrate strong correlation with thermal tolerance phenotypes in marine organisms [47].
In the field of visual ecology, understanding visual pigment function is crucial for deciphering how marine species perceive their environment. Microspectrophotometry (MSP) and opsin transcriptomics represent complementary approaches for visual pigment analysis, each with distinct strengths and limitations. MSP directly measures the spectral absorption properties of visual pigments in retinal photoreceptors, providing precise wavelength sensitivity data [50]. Opsin transcriptomics identifies and quantifies the expression of opsin genes encoding these visual pigments, offering insights into molecular genetic potential [51] [12].
This Application Note provides a detailed protocol for cross-validating these methodologies to achieve a comprehensive understanding of visual system function in marine species. The integrated approach allows researchers to correlate genetic capacity with functional expression, revealing mechanisms of visual adaptation to aquatic light environments.
Theoretical Framework and Principles
Visual Pigment Structure and Function
Visual pigments consist of an opsin protein bound to a chromophore (typically 11-cis-retinal). The spectral tuning of visual pigments—their specific wavelength absorption maxima (λmax)—is determined by amino acid substitutions in the opsin protein that alter the interaction with the chromophore [12]. Marine organisms inhabiting different depth zones or water types face distinct photic challenges, driving evolutionary adaptations in their visual systems through molecular changes to their opsin complements.
Methodological Synergies
MSP and transcriptomics offer complementary data streams. While MSP reveals the functional phenotype of visual pigments at a specific time point, opsin transcriptomics uncovers the genetic blueprint and expression patterns that determine visual potential [51]. Cross-validation strengthens conclusions by addressing methodological limitations: MSP cannot distinguish between highly similar opsin types, while transcriptomics alone cannot confirm whether expressed genes produce functional visual pigments. This integration is particularly valuable for studying marine species with complex visual systems adapted to specific photic environments.
Experimental Protocols
Microspectrophotometry (MSP) Workflow
Sample Preparation
- Retinal Tissue Isolation: Euthanize specimen following appropriate ethical guidelines. Enucleate eyes and rapidly dissect to isolate retinal tissue under dim red light to prevent pigment bleaching.
- Photoreceptor Suspension: Tease retinal tissue apart in chilled marine physiological saline or buffer (e.g., phosphate-buffered saline at appropriate osmolarity for marine species). Gently homogenize to create a photoreceptor suspension.
- Slide Preparation: Place small droplet of suspension between quartz microscope slide and coverslip. Seal edges to prevent evaporation during measurement [50].
MSP Measurement Procedure
- Instrument Calibration: Calibrate microspectrophotometer with neutral density filters and known standards. Ensure monochromator provides precise wavelength selection across relevant spectrum (typically 350-700 nm).
- Spectral Scanning: Identify individual photoreceptor outer segments under infrared illumination. Perform baseline scan outside cell. Scan through visual pigment spectrum with measuring beam positioned over outer segment.
- Bleaching Control: Expose visual pigment to bright white light for 1-3 minutes to bleach photopigment. Repeat scan to determine photoproduct absorption and validate pigment identity [50].
Data Analysis
- Absorbance Calculation: Subtract baseline and photoproduct spectra from sample scan to determine difference spectrum.
- λmax Determination: Fit data to visual pigment template (e.g., Govardovskii template) to determine wavelength of maximum absorption (λmax).
- Statistical Analysis: Compile measurements from multiple cells (typically 5-10 per photoreceptor type) to establish mean λmax values and variability [50].
Opsin Transcriptomics Workflow
RNA Extraction and Sequencing
- Retinal Tissue Preservation: Preserve retinal samples in RNAlater or flash-freeze in liquid nitrogen immediately after dissection to prevent RNA degradation.
- RNA Extraction: Homogenize tissue and extract total RNA using commercial kits (e.g., TRIzol method or column-based kits). Assess RNA quality and integrity using Bioanalyzer or similar (RIN > 8.0 recommended).
- Library Preparation and Sequencing: Prepare sequencing libraries using appropriate kits (e.g., Illumina TruSeq). For low-input samples, employ amplification methods. Sequence on appropriate platform (Illumina HiSeq/MiSeq recommended for good coverage) [51] [12].
Bioinformatics Analysis
- Transcriptome Assembly: De novo assemble sequenced reads into transcripts using assemblers like Trinity or SOAPdenovo-Trans if reference genome unavailable.
- Opsin Identification: Search assembled transcripts for opsin sequences using BLAST with known opsin sequences as queries. Identify complete open reading frames and check for absence of premature stop codons that would indicate pseudogenization.
- Expression Quantification: Map reads back to assembled transcripts to estimate expression levels (e.g., using RSEM or similar tools). Normalize reads to transcripts per million (TPM) or fragments per kilobase million (FPKM) [51] [12].
- Phylogenetic Analysis: Align identified opsin sequences with orthologs from related species. Construct phylogenetic trees to confirm opsin class identity and identify gene duplication events [12].
- Spectral Tuning Site Analysis: Translate nucleotide sequences to amino acid sequences. Identify known spectral tuning sites and compare with species of known λmax to predict spectral sensitivities [12].
Integrated Cross-Validation Workflow
The following diagram illustrates the strategic integration of MSP and transcriptomics methodologies throughout the experimental timeline:
Data Integration and Analysis
Comparative Analysis Framework
Effective integration requires systematic comparison of MSP and transcriptomics datasets. Create a reference table aligning identified opsin genes with measured photoreceptor classes:
Table 1: Cross-Method Data Alignment Framework
| Opsin Gene Identity | Predicted λmax (nm) | MSP-Measured λmax (nm) | Expression Level | Photoreceptor Class |
|---|---|---|---|---|
| LWS | 560-570 | 562.3 ± 3.2 | 45.2 TPM | Long-wave sensitive cone |
| RH2 | 480-530 | 505.6 ± 4.1 | 32.7 TPM | Medium-wave sensitive cone |
| SWS2 | 410-490 | 448.9 ± 5.3 | 28.1 TPM | Short-wave sensitive cone |
| RH1 (rod) | 480-510 | 498.2 ± 2.8 | 68.9 TPM | Rod photoreceptor |
Interpretation Guidelines
- Concordant Results: Agreement between methods provides strong evidence for functional visual capabilities. Example: Expression of LWS opsin with intact key tuning sites correlates with MSP measurement of LWS cone λmax ~560 nm [51].
- Discordant Results: Discrepancies require further investigation:
- MSP detects pigments not found in transcriptome: Consider incomplete transcriptome assembly or unannotated opsin genes.
- Opsin genes expressed but not detected by MSP: Consider low expression levels, non-retinal expression, or improper folding.
- λmax predictions mismatch measurements: Consider novel tuning mechanisms, alternative chromophores (A1/A2), or post-translational modifications [12].
Advanced Applications
The integrated approach enables investigation of complex visual adaptations:
Table 2: Marine Visual Adaptation Case Studies
| Adaptation Phenomenon | MSP Signature | Transcriptomic Signature | Marine Example |
|---|---|---|---|
| Deep-water spectral shift | Short-wavelength shifted λmax | Overexpression of SWS opsins | Deep-sea fishes [12] |
| Chromatic adaptation | Broaden absorbance spectra | Co-expression of multiple opsin types | Mesopelagic crustaceans [40] |
| Life stage differentiation | Different photoreceptor complements | Developmental shift in opsin expression | Diadromous species |
| A1/A2 chromophore mixing | Variable λmax measurements | Altered retinoid metabolism genes | Freshwater/marine transitions [12] |
The Scientist's Toolkit
Table 3: Essential Research Reagents and Solutions
| Item | Specification | Application | Notes |
|---|---|---|---|
| MSP Solutions | |||
| Marine physiological saline | Appropriate osmolarity for target species | Retinal dissection and photoreceptor suspension | Adjust for species-specific requirements |
| Transcriptomics Solutions | |||
| RNA stabilization reagent | RNAlater or similar | Tissue preservation for RNA extraction | Critical for field work in remote locations |
| RNA extraction kit | Column-based or TRIzol | Total RNA isolation from retinal tissue | Assess quality spectroscopically |
| cDNA synthesis kit | Reverse transcription | Library preparation for sequencing | Include controls without reverse transcriptase |
| Cross-Method Validation | |||
| Opsin reference sequences | Curated database | Bioinformatics identification | Compile from related species |
| Spectral template libraries | Standard visual pigment templates | λmax determination from MSP data | Govardovskii et al. templates commonly used |
Visualizing Molecular to Functional Relationships
The relationship between opsin gene expression, protein structure, and functional visual pigment measurement can be visualized as follows:
Integrating MSP with opsin transcriptomics provides a powerful cross-validation framework for visual pigment research in marine species. This approach moves beyond the limitations of single-method studies, enabling comprehensive understanding of visual adaptation mechanisms from gene sequence to functional phenotype. The protocols outlined here provide a standardized methodology applicable to diverse marine taxa, facilitating comparative studies of visual evolution across different aquatic light environments.
Ensuring Accuracy: MSP Validation and Comparative Analytical Techniques
Application Note: Principles of Inter-Laboratory Comparison for MSP Data Quality
Microspectrophotometry (MSP) is a sophisticated technique for measuring the absorbance spectra of visual pigments in single photoreceptor cells. In marine species research, where visual adaptations are critical for survival, the integrity of this data is paramount. The principle of inter-laboratory comparison serves as a formal process to establish confidence in measurement results by demonstrating that the uncertainty specifications of the calibration capabilities of participating laboratories are correct [52]. This practice is directly analogous to benchmarking in commercial sectors, where organizations like Managed Service Providers (MSPs) use standardized surveys and key performance indicators (KPIs) to gauge their operational performance against peers [53] [54]. For scientific research, applying a structured benchmarking approach ensures that MSP data for marine visual pigments is both reliable and comparable across different studies and institutions.
Core Concepts and Quantitative Benchmarks
The following table summarizes key quantitative benchmarks and their implications for MSP data quality, drawing parallels from established laboratory medicine practices [54].
Table 1: Key Benchmarking Metrics for Data Quality
| Metric Category | Current Benchmark | Implication for MSP Practice |
|---|---|---|
| Formal Quality Management | Variable adoption of standards (e.g., ISO 15189) | Implementation of a Quality Management System is crucial for patient safety and data reproducibility [54]. |
| KPI Monitoring Rate | 10-30% of laboratories overall | Active monitoring of instrument performance KPIs is essential for continuous improvement [54]. |
| Diagnosis & Treatment Speed | Only 19% of laboratories monitor related KPIs | Highlights a common gap; in MSP, this translates to tracking data throughput and analysis speed [54]. |
| Automation & Digitalization | Identified as a critical need | Reduces manual errors and improves efficiency in data collection and processing [54]. |
The primary statistical criterion for assessing a laboratory's results in a comparison is the normalized error (Eₙᵢ), where a value of ≤1 generally indicates a passing result. However, this criterion has limitations, as high values for the uncertainty of the transfer standard (uTS) or a participant's measurement repeatability (urepeat) can artificially lower the Eₙᵢ value, potentially allowing subpar performance to pass. A more robust assessment involves evaluating the comparison uncertainty (ucomp), defined as the root-sum-of-squares of uTS and u_repeat, which provides a better tool for gauging the stringency of the comparison itself [52].
Protocol: Implementing a Comparative Study for Marine Visual Pigment MSP
Experimental Workflow for Inter-Laboratory MSP Comparison
The diagram below outlines the standardized workflow for conducting an inter-laboratory comparison of MSP measurements on marine visual pigments. This process is designed to ensure data consistency and reliability across participating labs.
Detailed Methodology
Phase 1: Study Design and Preparation
- Define Measurands and Scope: The primary measurand for visual pigment studies is typically the wavelength of maximum absorbance (λmax) for pigments such as rhodopsin and metarhodopsin. The study should specify the target species or pigment type, for example, defining the expected λmax for rhodopsin from a species like Euphausia pacifica as approximately 483 nm [4].
- Select Participating Laboratories: Laboratories should be selected based on their expertise in MSP and their use of comparable, yet potentially diverse, instrumentation. This diversity tests the robustness of the standardized protocol.
- Prepare and Distribute Reference Material: A stable and well-characterized reference material is the cornerstone of a valid comparison. This could be a synthetic visual pigment standard or a stable, homogenized biological sample. The reference material acts as the transfer standard, and its inherent uncertainty (u_TS) must be accurately quantified, as this value directly impacts the comparison's statistical power [52].
Phase 2: Standardized Measurement Protocol
All participating laboratories must adhere to a single, detailed MSP protocol to minimize inter-lab variability.
- Sample Preparation: For marine species, this involves isolating photoreceptors (e.g., rhabdoms) in a saline solution appropriate for the organism. The protocol must specify dissection techniques, solution pH, and temperature controls to prevent pigment degradation [4] [55].
- Instrument Calibration: The microspectrophotometer must be calibrated for wavelength accuracy using a holmium oxide filter or similar standard. Photometric accuracy should be verified with neutral density filters.
- Data Collection Parameters:
- Spectral Range: Typically 350-650 nm.
- Bandwidth: Set to 1-2 nm for sufficient resolution.
- Number of Scans: A minimum of 10 scans per sample should be averaged to improve the signal-to-noise ratio.
- Baseline Correction: A baseline scan with a clear path must be performed and subtracted from all sample measurements.
- Data Analysis: The λmax must be determined from the mean absorbance spectrum after smoothing, using a digital least squares algorithm to reduce noise [5]. The protocol should explicitly forbid manual estimation from chart recordings.
Phase 3: Data Analysis and Benchmarking
- Uncertainty Calculation: Each laboratory must report its measured λmax along with a comprehensive uncertainty budget. This includes Type A (statistical analysis of repeated measurements) and Type B (all other non-statistical sources) uncertainties, culminating in a combined standard uncertainty for its result [52].
- Calculate Normalized Error: For each laboratory
i, the normalized error is calculated as:Eₙᵢ = (LabResultᵢ - ReferenceValue) / √(uLabᵢ² + uRef²)
Whereu_Labis the combined standard uncertainty reported by the lab, andu_Refis the uncertainty of the reference value. An |Eₙᵢ| ≤ 1 indicates a passing result [52]. - Analysis and Reporting: The final report should present the results from all laboratories, highlight any outliers, and provide a statistical summary of the performance. As proposed in recent studies, using a probability-based criterion alongside Eₙ can help identify instances where results are "inconclusive" due to a high comparison uncertainty (u_comp), thus strengthening the conclusions drawn [52].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents and Materials for Visual Pigment MSP
| Item | Function / Explanation | Example from Literature |
|---|---|---|
| Multichannel Microspectrophotometer | Replaces photomultiplier with an array detector; eliminates mechanical scanning artifacts for more stable ship-based measurements [4]. | Core instrument described for sea-based studies on Euphausia pacifica [4]. |
| Isolated Rhabdom Preparations | The biological sample containing the visual pigments of interest; isolated from the retina of the marine organism. | Rhabdoms from Euphausia pacifica and deep-sea fish rods are the standard samples for analysis [4] [55]. |
| Physiological Saline Solution | A buffered salt solution to maintain the structural integrity and photochemical state of the isolated photoreceptors during analysis. | Implied as a standard for handling live tissue preparations from marine crustaceans and fish [4] [55]. |
| Wavelength Calibration Standards | Materials with known, sharp absorption peaks used to verify the wavelength accuracy of the microspectrophotometer. | Holmium oxide filters are a common industry standard for wavelength verification. |
| Neutral Density Filters | Filters that attenuate light uniformly across wavelengths; used to verify the photometric linearity and accuracy of the instrument. | Critical for ensuring absorbance measurements are quantitatively correct. |
| Digital Smoothing Algorithms | Software algorithms applied to spectral data to reduce high-frequency noise, allowing for more accurate determination of λmax. | Digital least squares smoothing of spectra is a standard data processing step [5]. |
Within visual ecology, a primary challenge is to fully describe an organism's visual capability by linking its genetic potential to its functional phenotype. The wavelength of maximum absorbance (λmax) of a visual pigment, determined via Microspectrophotometry (MSP), represents the functional phenotype. In contrast, genomic or transcriptomic analysis reveals the complement of opsin genes, the proteins that constitute the visual pigment, representing the genetic potential. This Application Note details the methodologies for correlating these two datasets, using examples from marine fish, particularly flatfish, to highlight the necessity of an integrated approach. A purely genomic inventory can be misleading, as the expressed opsin repertoire and the resulting visual pigments are dynamically tuned through development and in response to the environment [2] [56].
Theoretical Background: Visual Pigment Composition and Spectral Tuning
Visual pigments are composed of an opsin protein bound to a chromophore, either 11-cis retinal (A1) or 11-cis 3,4-dehydroretinal (A2). The λmax is determined by specific interactions between the opsin and the chromophore.
- Opsin Spectral Tuning: Amino acid substitutions at key sites within the opsin's chromophore-binding pocket can shift the λmax by altering the energy required to excite the chromophore [12]. For instance, the evolution of green sensitivity in a duplicated LWS opsin in Characiformes was achieved through substitutions at key tuning sites [12].
- Chromophore Shifts: The A2 chromophore produces visual pigments that are red-shifted by ~20–60 nm compared to A1-based pigments [13]. The ratio of A1/A2 chromophores can be modulated by environmental factors such as light regime and temperature [13].
- Novel Tuning Mechanisms: Recent work on coral anthozoan-specific opsins (ASO-II) has revealed a novel mechanism where a chloride ion (Cl⁻) acts as the counterion to stabilize the protonated Schiff base, a role fulfilled by a negatively charged amino acid in most other animal opsins [57].
Quantitative Data from Model Marine Species
The following table summarizes key findings from integrated studies on marine species, demonstrating the complex relationship between genomic repertoire and measured visual pigments.
Table 1: Correlated Opsin and Visual Pigment Data in Select Marine Fishes
| Species | Analytical Method | Opsin Repertoire (Genomic/Transcriptomic) | Measured Visual Pigments (λmax in nm) | Key Discrepancy/Interpretation |
|---|---|---|---|---|
| Atlantic Halibut (Hippoglossus hippoglossus) | MSP & Bioinformatics [2] | Detected via genome query | Rods: 491Cones: 431, 457, 500, 514, 527, 550 | MSP identified six cone visual pigments that only partially matched the opsin repertoire detected bioinformatically. |
| Starry Flounder (Platichthys stellatus) | MSP & Digital PCR [56] | Seven cone opsins: SWS1, SWS2B, SWS2A2, SWS2A1, RH2A1, RH2A2, LWS | Rods: 507 (young) → 517 (large)Cones: 437→456, 527→545 (with intermediates) | Visual pigment λmax changes with growth due to opsin switches and co-expression, e.g., S(445) and M(536) pigments had widened bandwidths, indicating opsin co-expression. |
| Masked Greenling (Hexagrammos octogrammus) | MSP & HPLC [13] | Not specified | A1/A2 pigment mixtures in rods and cones; λmax varied with light adaptation. | Light adaptation induced an unusual A1→A2 conversion, opposite the common pattern, leading to spectral shifts. |
Detailed Experimental Protocols
Protocol A: Microspectrophotometry (MSP) for λmax Determination
MSP directly measures the absorbance spectrum of single photoreceptor cells, providing the definitive λmax value for visual pigments present in the retina.
4.1.1 Sample Preparation
- Dark Adaptation: Euthanize the fish and dissect the retina under dim red light or in complete darkness to allow visual pigments to fully regenerate.
- Retinal Isolation: Isolate the retina in a physiological saline solution.
- Photoreceptor Dissociation: Gently macerate a small piece of retina between a microscope slide and coverslip to release individual photoreceptors.
4.1.2 Data Collection & Analysis
- Instrumentation: Utilize a single-beam or multichannel MSP. The latter eliminates scanning-related artifacts and is suitable for ship-based studies on fragile marine organisms [4].
- Measurement: Select a single rod or cone outer segment. A baseline transmission spectrum (I₀) is taken, followed by a sample spectrum (I). Absorbance is calculated as log(I₀/I).
- Bleaching Control: Expose the photoreceptor to intense broadband light to bleach the visual pigment. A subsequent measurement confirms the loss of the pigment's absorbance band.
- Template Fitting: The pre-bleach absorbance spectrum is fitted to standard visual pigment templates (e.g., Govardovskii et al., 2000) for A1 or A2 chromophores to determine the λmax [13] [56]. A widened bandwidth often suggests co-expression of multiple opsins within a single photoreceptor [56].
Figure 1: MSP Workflow
Protocol B: Genomic and Transcriptomic Analysis of Opsin Repertoire
This protocol identifies the full set of opsin genes and their expression.
- DNA/RNA Extraction: Extract high-quality genomic DNA and/or retinal mRNA.
- Sequencing and Assembly: Perform whole-genome sequencing and/or retinal transcriptome (RNA-Seq) sequencing. De novo or reference-guided assembly is used to reconstruct sequences.
- Opsin Identification: Use homology search tools (e.g., BLAST) against known opsin sequences to identify candidate genes in the genome or transcriptome.
- Sequence Analysis:
- Phylogenetic Analysis: Classify opsins into correct subfamilies (SWS1, SWS2, RH2, LWS, RH1).
- Tuning Site Analysis: Compare amino acid sequences at known spectral tuning sites to predict approximate λmax shifts [12].
- Gene Expression: For RNA-Seq data, calculate normalized read counts (e.g., FPKM or TPM) to determine the relative expression level of each opsin.
Protocol C: Correlative Analysis and Validation
This critical step integrates data from MSP and genomic analyses.
- Reconcile Numbers: Compare the number of distinct visual pigment λmax values from MSP with the number of expressed opsin genes from transcriptomics.
- Identify Co-expression: If MSP reveals visual pigments with widened bandwidths or intermediate λmax values not matching a single opsin template, this is strong evidence for co-expression of two opsin types in a single photoreceptor cell [56]. The contribution of each opsin can be quantified by selective bleaching [56].
- Account for Chromophore: Determine the A1:A2 ratio in retinal extracts using High-Performance Liquid Chromatography (HPLC) [13] [56]. This explains discrepancies between the λmax predicted for an A1-based opsin and the measured value if an A2 chromophore is present.
- In situ Hybridization (ISH): Validate opsin co-expression and mosaic organization by using fluorescently labeled RNA probes to localize specific opsin mRNAs within retinal sections [2].
Figure 2: Data Correlation Strategy
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Reagents for Visual Pigment Research
| Category | Item | Function/Application |
|---|---|---|
| Sample Preparation | Physiological Saline | Maintains osmotic balance and health of retinal tissue during dissection. |
| 9-cis or 11-cis Retinal | Chromophore used to regenerate visual pigments in heterologous expression systems or reconstitute pigments in vitro [58] [57]. | |
| Molecular Biology | RNA/DNA Extraction Kits | Isolate high-quality nucleic acids for sequencing. |
| Reverse Transcriptase, PCR Kits | For cDNA synthesis and amplification of opsin genes. | |
| In situ Hybridization Kits | For cellular localization of opsin mRNA transcripts in retinal sections. | |
| Biochemistry | HPLC System with UV/Vis Detector | To analyze and quantify the A1/A2 chromophore ratio in retinal extracts [13]. |
| Detergent (e.g., n-Dodecyl β-D-maltoside) | Solubilizes opsin proteins from cell membranes for purification and spectroscopic analysis [57]. | |
| Cell-Based Assays | HEK293 or COS-1 Cell Lines | Heterologous expression systems for characterizing the spectral properties and G-protein coupling of cloned opsin genes [58] [57]. |
| Luminescent Reporters (e.g., GloSensor, Aequorin) | Measure second messenger responses (cAMP, Ca²⁺) to light activation in cell-based assays [58]. |
Concluding Remarks
Correlating MSP-derived λmax with the genomic opsin repertoire is not a simple one-to-one mapping. It is a sophisticated process that reveals the dynamic plasticity of the visual system. As demonstrated in marine fishes, organisms actively tune their visual capabilities through opsin switches, co-expression, and chromophore variation [2] [13] [56]. Relying solely on genomic data risks overlooking these critical regulatory mechanisms. Therefore, MSP remains the gold standard for functional validation, and its integration with modern genomic tools is essential for a complete understanding of visual ecology and evolution.
This application note provides a comparative framework for Microspectrophotometry (MSP), Fourier-Transform Infrared (FTIR) Spectroscopy, and Raman Spectroscopy within marine visual pigment research. These techniques enable researchers to investigate the molecular basis of vision in marine species, from the spectral tuning of visual pigments to the characterization of fluorescent body patterns. The selection of an appropriate technique is critical for addressing specific research questions about the structure, function, and ecology of visual systems in the marine environment. This document outlines detailed protocols, provides a comparative analysis of technical specifications, and recommends essential research reagents to guide experimental design.
Experimental Protocols
Protocol for Microspectrophotometry (MSP) of Marine Fish Retinas
MSP is used to characterize the spectral absorbance of visual pigments within individual photoreceptor cells [13] [59].
- Sample Preparation: Dark-adapt animals for a minimum of 12 hours. Under dim red light, enucleate the eyes and hemisect. Remove the retina and immerse in a suitable physiological saline or tissue culture medium. Gently tease apart small pieces of retina on a coverslip to isolate photoreceptor cells [13].
- Equipment Setup: Utilize a microspectrophotometer equipped with a bright monochromatic light source and a high-sensitivity detector. Calibrate the system using known spectral standards and a blank area of the preparation slide.
- Data Acquisition: Select intact rod and cone outer segments for measurement. Record a baseline scan (I0). Scan the cell across the relevant wavelength range (e.g., 350-750 nm) to obtain the sample transmission spectrum (I). A second baseline scan should be performed after the sample scan to confirm stability.
- Data Processing: Calculate absorbance as A = log10(I0/I). Fit the resulting absorbance spectrum with standard visual pigment templates to determine the wavelength of maximum absorbance (λmax). For pigments containing a mixture of chromophores, the data may be best fitted by a combination of A1- and A2-based visual pigment templates [13].
Protocol for ATR-FTIR Spectroscopy of Biological Samples
ATR-FTIR spectroscopy provides a rapid, non-destructive snapshot of the overall molecular composition of a sample and is useful for monitoring biochemical changes [60] [61].
- Sample Preparation: For tissue or biomass (e.g., seaweed, fish fillets), freeze-dry and homogenize into a fine powder using a mortar and pestle or ball mill. For liquid samples, a drying step may be required unless using a liquid ATR accessory [62] [61].
- Equipment Setup: Use an FTIR spectrometer equipped with an ATR crystal. Clean the crystal surface with a suitable solvent and perform a background scan with a clean, empty ATR.
- Data Acquisition: Place a small amount of the prepared solid sample onto the ATR crystal and ensure good contact by applying uniform pressure. Acquire spectra typically over the mid-infrared range (e.g., 4000–400 cm⁻¹) with a resolution of 4 cm⁻¹. Accumulate multiple scans to improve the signal-to-noise ratio [60].
- Data Processing: Perform atmospheric suppression and baseline correction. For complex biological matrices, multivariate analysis (e.g., Principal Component Analysis (PCA) or Partial Least Squares Regression (PLSR)) is essential for interpreting spectral variance and developing quantitative models for specific compounds [60] [61].
Protocol for Raman Spectroscopy of Pigments and Tissues
Raman spectroscopy is highly effective for identifying specific molecular structures, such as pigments, based on their vibrational fingerprints [63] [64].
- Sample Preparation: Samples can be analyzed in situ with minimal preparation. For pigment identification, a pure sample can be placed on a glass slide or aluminum foil. For retinal visual pigments, intact photoreceptors may be frozen on a cryostage for resonance Raman studies [63] [64].
- Equipment Setup: A Raman spectrometer with a microscope is standard. Select an appropriate laser excitation wavelength; 785 nm is common for reducing fluorescence, while shorter wavelengths may be used for resonance Raman experiments. Calibrate the instrument using a silicon standard.
- Data Acquisition: Focus the laser on the area of interest. Set acquisition time and number of accumulations to optimize signal while preventing sample degradation. Acquire spectra over the desired Raman shift range (e.g., 100–3300 cm⁻¹) [63] [65].
- Data Processing: Apply a background correction algorithm to subtract native fluorescence and any cosmic rays. Compare the resulting spectrum against reference spectral libraries for positive identification of molecular components [63].
Technical Comparison and Data Presentation
The table below summarizes the core principles, applications, and key specifications of the three analytical techniques.
Table 1: Technical Comparison of MSP, FTIR, and Raman Spectroscopy
| Feature | Microspectrophotometry (MSP) | Fourier-Transform Infrared (FTIR) Spectroscopy | Raman Spectroscopy |
|---|---|---|---|
| Core Principle | Measures absorbance of light by single cells | Measures absorption of IR light by molecular bonds | Measures inelastic scattering of light by molecular bonds |
| Primary Application in Marine Research | Spectral sensitivity of photoreceptors; visual pigment λmax determination [13] [59] | Biochemical profiling of tissues; rapid quality assessment; metabolic status [60] [61] | Pigment identification; molecular structure of pigments and polymers [63] [64] |
| Spectral Range | UV-Visible (e.g., ~350-750 nm) | Mid-IR (e.g., 4000-400 cm⁻¹) | Raman Shift (e.g., 100-3300 cm⁻¹) |
| Spatial Resolution | High (single cell) | Low to Moderate (bulk powder or tissue area) | High (sub-micron with microscope) |
| Key Output | Absorbance spectrum, λmax | Absorption spectrum, functional group identification | Raman shift spectrum, molecular fingerprint |
| Sample Form | Intact photoreceptor cells in solution | Dried powder, solid tissue, liquids (with accessories) | Solids, liquids, gases; minimal preparation |
| Quantitative Capability | Direct quantification of λmax and pigment density | Excellent with multivariate calibration (PLSR) [60] [61] | Good with established calibration models |
Table 2: Representative Quantitative Data from Spectroscopy Studies
| Study Focus | Technique | Key Quantitative Finding | Statistical Performance |
|---|---|---|---|
| Gilthead Sea Bream Quality [60] | FTIR + PLS-R | Prediction of Total Viable Counts (TVC) | R² = 0.98, RMSEC = 0.43 log CFU/g (Aerobic) [60] |
| Ulva Seaweed Composition [61] | ATR-FTIR + PLS-R | Prediction of carbohydrate concentration | Effective model developed [61] |
| Marine Microplastics [66] | Raman vs. FTIR | MP count (≤500 μm) | Raman detected 2x higher MP numbers than FTIR [66] |
| Fairy Wrasse Visual Pigments [59] | MSP | λmax of cone photoreceptors | Single: 514 nm; Twin A: 498 nm; Twin B: 532 nm [59] |
Experimental Workflow and Logical Relationships
The following diagram illustrates a decision-making workflow for selecting and applying these techniques within a marine biology research context.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for Spectroscopy in Marine Research
| Item | Function/Application | Example Use Case |
|---|---|---|
| Physiological Saline | Maintains viability and osmotic balance of retinal tissue during dissection and MSP analysis. | Isolation of photoreceptor cells from marine fish retinas [13]. |
| Visual Pigment Templates (A1/A2) | Mathematical models used to fit MSP absorbance data and determine the wavelength of maximum absorption (λmax). | Differentiating between rhodopsin and porphyropsin pigments in fish photoreceptors [13]. |
| Plate Count Agar (PCA) | Growth medium for enumerating microbial populations (e.g., Total Viable Counts). | Providing reference data for correlating microbial spoilage with FTIR spectral data in fish [60]. |
| Gold-Coated Filters | Substrate for concentrating and analyzing microplastic particles via automated Raman or FTIR imaging. | Analysis of microplastics ≤500 μm in aquatic environmental samples [66]. |
| ATR Crystal (ZnSe, Diamond) | Internal reflection element in ATR-FTIR spectroscopy that contacts the sample for infrared measurement. | Analyzing the biochemical composition of dried and powdered seaweed or fish tissue [60] [61]. |
| Silicon Wafer Standard | Reference material for calibrating the wavelength and intensity of a Raman spectrometer. | Ensuring accurate and reproducible Raman spectral measurements of pigments or polymers [63]. |
Abstract Establishing a visual function for biofluorescence in marine species requires moving beyond observational accounts to a hypothesis-driven, criteria-based framework. This Application Note details a four-criteria protocol for determining if fluorescence creates a meaningful visual signal under natural conditions. The protocols are contextualized within microspectrophotometry (MSP) research, providing methodologies for quantifying visual pigment absorbance, fluorescent emissions, and the contribution of fluorescence to an organism's overall optical signature. This framework is essential for validating the numerous proposed ecological roles of fluorescence, from communication to camouflage.
The discovery of green fluorescent protein (GFP) in the jellyfish Aequorea victoria ignited a scientific revolution in cell biology and has since led to the recognition of widespread biofluorescence across marine lineages, including fishes and corals [67]. In marine environments, where ambient light becomes increasingly monochromatic and blue-shifted with depth, fluorescence is hypothesized to serve critical visual functions such as intraspecific signaling, prey attraction, and camouflage [67] [68]. However, the compelling appearance of fluorescence under artificial blue-light excitation does not, in itself, constitute evidence of a biological function [69]. A rigorous, multi-step framework is necessary to distinguish adaptive visual signals from evolutionarily neutral epiphenomena. This document outlines the requisite protocols and experimental controls to define a visual function for fluorescence, with a specific focus on the use of microspectrophotometry in marine species research.
A Four-Criteria Framework for Establishing Visual Function
A robust demonstration of a visual function for fluorescence must satisfy four key criteria [69]. The following sections detail experimental protocols for each.
Criterion 1: Establish the Phenomenon in the Natural Context Objective: Confirm that fluorescence occurs under illumination from wavelengths naturally available in the organism's habitat.
Protocol 1.1: In Situ Fluorescence Imaging
- Excitation Source: In field or lab settings, illuminate the subject with a narrow-band blue light source (e.g., 440–460 nm or 490 ±5 nm) to mimic dominant ambient wavelengths in marine environments [70] [69].
- Emission Filtering: Use a long-pass emission filter (e.g., 514 nm or 561 nm LP) on the camera lens to block reflected excitation light and isolate the emitted fluorescence [70].
- Documentation: Capture images and videos to record the spatial patterning and intensity of the fluorescence.
Criterion 2: Confirm Signal Reception Capability Objective: Verify that the intended receiver (e.g., a conspecific) possesses the physiological capacity to detect the fluorescent emission.
Protocol 2.1: Microspectrophotometry (MSP) for Visual Pigment Analysis This protocol is core to determining the spectral sensitivity of the receiver's retina.
- Tissue Preparation: Under dim red light, dissect the retina and prepare photoreceptor cells for analysis. Isolate outer segments in a suitable saline solution [2] [12].
- Instrumentation: Utilize a single- or double-beam microspectrophotometer. A multichannel instrument can enhance the signal-to-noise ratio [40].
- Spectral Absorbance Measurement:
- Focus a monochromatic measuring beam onto a single photoreceptor outer segment.
- Scan through a range of wavelengths (e.g., 350–650 nm) to obtain an absorbance spectrum.
- Determine the wavelength of maximum absorbance (λmax) for each visual pigment. For example, MSP has revealed six cone visual pigments in Atlantic halibut, with λmax values ranging from 431 nm to 550 nm [2].
- Data Analysis: Fit absorbance data to a visual pigment template to confirm λmax. Compare the λmax of the receiver's visual pigments to the emission spectra of the fluorescence (see Protocol 3.2) to assess potential detectability.
Protocol 2.2: Intraocular Filter Transmission
- Sample Extraction: Carefully extract the lens and other potential ocular filters.
- Spectrometry: Use a conventional spectrometer to measure the transmission spectrum of the filter.
- Analysis: Determine if the filter acts as a long-pass filter, which could enhance the perception of longer-wavelength fluorescence in a blue-dominated environment [68].
Criterion 3: Quantify the Contribution to Optical Signature Objective: Model and measure whether fluorescence contributes meaningfully to the subject's appearance under relevant ambient illumination.
Protocol 3.1: Measuring Reflectance and Fluorescence Efficiency
- Setup: Illuminate a small area of the subject's skin with monochromatic light and measure the full optical output with a spectrometer.
- Separate Signals: The total optical signature is the sum of reflected light and fluoresced light. The key metric is the fluorescence efficiency—the proportion of incident photons that are converted and emitted as fluorescence [69].
- Calculation: A fluorescence efficiency of >1% is a suggested benchmark for a potentially meaningful contribution, though this is context-dependent [69].
Protocol 3.2: Fluorescent Emission Spectrometry
- Excitation: Use the same blue excitation light as in Protocol 1.1.
- Measurement: Position a fiber optic probe connected to a portable spectrophotometer (e.g., Ocean Optics USB2000+) close to the fluorescent tissue.
- Spectral Recording: Record the emission spectrum, identifying the peak emission wavelength(s). Document the remarkable diversity; for instance, marine teleosts can exhibit multiple distinct emission peaks within the green and red spectrum, from ~500 nm to beyond 650 nm [70].
Criterion 4: Demonstrate a Behavioral Response Objective: Provide conclusive evidence that the viewing organism's behavior is influenced by the fluorescence.
Protocol 4.1: Controlled Behavioral Experiments
- Experimental Design: Create a two-choice setup where test subjects can interact with stimuli under controlled lighting.
- Stimuli: Use realistic models or video playback of conspecifics with and without their natural fluorescent patterning.
- Lighting Control: Illuminate the setup with broad-spectrum "white" light that includes the natural excitation wavelengths for fluorescence. Crucially, run identical trials under lighting that lacks the excitation wavelengths (e.g., using a long-pass filter at the light source), thereby removing the fluorescence without altering other visual cues [69].
- Metrics: Quantify behavioral responses such as association time, aggression, or mating displays. A statistically significant difference in behavior between the two lighting conditions provides strong evidence for a visual function.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 1: Key reagents, materials, and instrumentation for fluorescence and visual ecology research.
| Item | Function/Application | Example Specifications |
|---|---|---|
| Narrow-band Blue LED Light | Provides excitation light to elicit fluorescence. | 490 nm ±5 nm interference filter [70]. |
| Long-Pass Emission Filters | Blocks reflected blue light, allowing only fluorescence to be imaged or measured. | 514 nm LP, 561 nm LP [70]. |
| Portable Spectrophotometer | Measures fluorescent emission spectra from specimens in lab or field. | Ocean Optics USB2000+ with fiber optic probe [70]. |
| Microspectrophotometer | Measures absorbance spectra of single photoreceptor cells to determine spectral sensitivity. | Single or double-beam design; multichannel for improved SNR [40]. |
| Opsin Gene Probes | For in-situ hybridization to localize opsin mRNA expression in retinal sections [2]. | Riboprobes against sws2, rh2, lws, etc. |
| A1/A2 Chromophores | The light-sensitive chromophores (11-cis retinal variants) that, bound to opsin, determine (\lambda_{max}) shifts [12]. | 11-cis retinal (A1); 11-cis-3,4-dehydroretinal (A2). |
Experimental Workflow: From Detection to Validation
The following diagram illustrates the logical and experimental progression through the four-criteria framework, integrating the protocols described above.
Experimental Workflow for Validating Visual Fluorescence
The proposed four-criteria framework provides a rigorous, repeatable path for testing hypotheses regarding the visual functions of biofluorescence. By systematically integrating imaging, microspectrophotometry, spectrometry, and behavioral assays, researchers can move beyond correlation to causation. This approach is critical for elucidating the complex interplay between fluorescent signals, visual physiology, and ecology in marine environments.
Conclusion
Microspectrophotometry stands as an indispensable, validated tool for deciphering the visual ecology of marine species, from mapping complex photoreceptor mosaics in flatfish to characterizing rare visual pigments in deep-sea organisms. The synergy between MSP and modern omics technologies creates a powerful pipeline for validating visual function and uncovering the genetic blueprints of light detection. These detailed insights into marine visual systems not only advance sensory ecology but also open innovative avenues for biomedical research. The principles of light capture and signal transduction studied in marine models can inspire novel approaches in optogenetics and imaging, while the organisms themselves remain a prolific source of uncharacterized bioactive compounds, underscoring the continued value of MSP in future marine bioprospecting and drug discovery initiatives.
References
- 1. Visual Pigment - an overview [https://www.sciencedirect.com/topics/earth-and-planetary-sciences/visual-pigment]
- 2. Photoreceptor distributions, visual pigments and the opsin ... [https://www.nature.com/articles/s41598-022-11998-9]
- 3. Spectral characteristics of visual pigments in two mullet ... [https://pubmed.ncbi.nlm.nih.gov/41058234/]
- 4. A multichannel microspectrophotometer for visual pigment ... [https://pubmed.ncbi.nlm.nih.gov/3660659/]
- 5. A multichannel microspectrophotometer for visual pigment ... [https://www.sciencedirect.com/science/article/pii/0042698987900198]
- 6. Oil droplets are severely overlooked in risk assessments [https://www.nature.com/articles/s43247-025-02805-0]
- 7. Retinal Structure of Poecilia sphenops: Photoreceptor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10967514/]
- 8. Scientists Thought This Creature Had No Brain. It's All Brain. [https://www.popularmechanics.com/science/animals/a69443677/all-brain-creature/]
- 9. Marine biologists discover 4 new types of photoreceptor [https://bigthink.com/hard-science/marine-organism-photoreceptors/]
- 10. Water Structure and Electric Fields at the Interface of Oil Droplets [https://pmc.ncbi.nlm.nih.gov/articles/PMC12335212/]
- 11. Step-by-step preparation of mouse eye sections for routine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8585658/]
- 12. Visual pigment evolution in Characiformes [https://pmc.ncbi.nlm.nih.gov/articles/PMC7451407/]
- 13. Unusual A1/A2–visual pigment conversion during light ... [https://www.sciencedirect.com/science/article/abs/pii/S1095643319303241]
- 14. Characterization of visual pigments, oil droplets, lens and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4495465/]
- 15. The evaluation of evidence for microspectrophotometry ... [https://www.sciencedirect.com/science/article/abs/pii/S0379073819304190]
- 16. Spectral absorption of visual pigments in stomatopod larval ... [https://link.springer.com/article/10.1007/s00359-015-1063-y]
- 17. Microspectrophotometry of single rhabdoms in the retina ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2228657/]
- 18. of Microspectrophotometry and oil droplets in... visual pigments [https://pubmed.ncbi.nlm.nih.gov/14978063/]
- 19. (PDF) Analysis of visual absorbance and... - Academia.edu pigment [https://www.academia.edu/10263533/Analysis_of_visual_pigment_absorbance_and_luminescence_emission_spectra_in_marine_ostracodes_Crustacea_Ostracoda]
- 20. Vibration Analysis of Typical Flat Plate Ship Structures ... [https://www.mdpi.com/2077-1312/13/1/57]
- 21. ABS publishes ship vibration analysis research [https://smartmaritimenetwork.com/2022/07/15/abs-publishes-ship-vibration-analysis-research/]
- 22. Vibration Monitoring of Rotating Machinery in Ships [https://www.vikinganalytics.se/blog/vibration-monitoring-of-rotating-machinery-in-ships-an-ai4eu-project]
- 23. of Microspectrophotometry and oil droplets in... visual pigments [https://researchers.mq.edu.au/en/publications/microspectrophotometry-of-visual-pigments-and-oil-droplets-in-a-m]
- 24. Adaptations of Cetacean Retinal Pigments to Aquatic ... [https://repository.library.noaa.gov/view/noaa/23798]
- 25. Parallel Spectral Tuning of a Cone Visual Pigment Provides ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11503649/]
- 26. of Baltic Sea fishes of Visual and limnic origin pigments marine [https://researchportal.helsinki.fi/en/publications/visual-pigments-of-baltic-sea-fishes-of-marine-and-limnic-origin]
- 27. Photoreceptor Differentiation during Retinal Development ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6730367/]
- 28. Avoidance of different durations, colours and intensities ... [https://www.nature.com/articles/s41598-021-97986-x]
- 29. Bird colour vision – from cones to perception [https://www.sciencedirect.com/science/article/abs/pii/S235215461930021X]
- 30. A detailed investigation of the visual system and ... [https://www.nature.com/articles/s41598-019-52297-0]
- 31. A Comprehensive Microscopy Analysis of the Retina ... [https://www.mdpi.com/2673-6004/6/1/7]
- 32. Visual adaptation of opsin genes to the aquatic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7690139/]
- 33. On visual pigment templates and the spectral shape of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2962788/]
- 34. Simple exponential functions describing the absorbance ... [https://www.sciencedirect.com/science/article/pii/004269899390237Q]
- 35. Microspectrophotometry - an overview [https://www.sciencedirect.com/topics/medicine-and-dentistry/microspectrophotometry]
- 36. Spontaneous activation of visual pigments in relation to ... [https://elifesciences.org/articles/18492]
- 37. Visual pigments of Baltic Sea fishes of marine and limnic ... [https://pubmed.ncbi.nlm.nih.gov/17822578/]
- 38. Physiological studies of the interaction between opsin and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3561676/]
- 39. Microspectrophotometry [https://www.microtrace.com/technique/microspectrophotometry/]
- 40. A multichannel microspectrophotometer for visual pigment ... [https://www.sciencedirect.com/science/article/abs/pii/0042698987900198]
- 41. Visual Pigments of Crayfish [https://www.nature.com/articles/2151131a0]
- 42. The Assessment of Visual Function and Functional Vision [https://pmc.ncbi.nlm.nih.gov/articles/PMC6761988/]
- 43. Scientific Data Presentation: a Picture Is Worth a Thousand ... [https://www.aje.com/arc/scientific-data-presentation-picture-worth-thousand-words]
- 44. Presenting data in tables and charts - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4008059/]
- 45. Elements must meet minimum color contrast ratio thresholds [https://dequeuniversity.com/rules/axe/4.8/color-contrast]
- 46. Marine probiotics: increasing coral resistance to bleaching ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6461899/]
- 47. Intergenerational metabolomic signatures of bleaching ... [https://www.nature.com/articles/s41467-025-61102-8]
- 48. Applications of visible spectral imaging technology for ... [https://www.nature.com/articles/s40494-024-01434-8]
- 49. Estimation of the age of human bloodstains under the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5643403/]
- 50. Intraspecific geographic variation in rod and cone visual ... [https://www.nature.com/articles/srep41445]
- 51. Caecilians maintain a functional long-wavelength-sensitive ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11839130/]
- 52. Evaluating Inter-Laboratory Comparison Data [https://www.nist.gov/publications/evaluating-inter-laboratory-comparison-data]
- 53. State of the MSP Industry Report [https://www.mspglobal.com/state-of-the-industry-report/]
- 54. Benchmarking medical laboratory performance on a global ... [https://pubmed.ncbi.nlm.nih.gov/38952740/]
- 55. Visual pigments in the individual rods of deep-sea fishes [https://link.springer.com/article/10.1007/BF00612519]
- 56. Parallel opsin switches in multiple cone types of the starry flounder retina: tuning visual pigment composition for a demersal life style | Scientific Reports [https://www.nature.com/articles/s41598-018-23008-y]
- 57. Coral anthozoan-specific opsins employ a novel chloride ... [https://elifesciences.org/articles/105451]
- 58. Human melanopsin forms a pigment maximally sensitive to blue light (λmax ≈ 479 nm) supporting activation of Gq/11 and Gi/o signalling cascades - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3619500/]
- 59. Fluorescence characterisation and visual ecology of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4791940/]
- 60. Application of Fourier Transform Infrared (FT-IR) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9367857/]
- 61. ATR-FTIR spectroscopy combined with multivariate ... [https://www.frontiersin.org/journals/marine-science/articles/10.3389/fmars.2023.1154461/full]
- 62. Infrared and Raman spectroscopy of blood plasma for rapid ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12304263/]
- 63. Raman Spectroscopy for Pigment Identification [https://www.spectroscopyonline.com/view/raman-spectroscopy-pigment-identification]
- 64. Why are blue visual pigments blue? A resonance Raman ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC286728/]
- 65. Raman Spectroscopy of Marine Microplastics [https://www.sciencedirect.com/science/article/abs/pii/S0141113621000696]
- 66. Comparison of Raman and Fourier Transform Infrared ... [https://pubmed.ncbi.nlm.nih.gov/30350953/]
- 67. Sea as a color palette: the ecology and evolution of fluorescence [https://pmc.ncbi.nlm.nih.gov/articles/PMC7288533/]
- 68. Repeated and widespread evolution of biofluorescence in ... [https://www.nature.com/articles/s41467-025-59843-7]
- 69. Method for Determining the Contribution of Fluorescence to ... [https://www.frontiersin.org/journals/marine-science/articles/10.3389/fmars.2017.00266/full]
- 70. Marine fishes exhibit exceptional variation in biofluorescent ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0316789]